Helena Rasche

Favourite Topics
Favourite Formats
Contributions
The following list includes only slides and tutorials where the individual has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
GitHub Activity
Tutorials
- Genome Annotation / Refining Genome Annotations with Apollo (prokaryotes)
- Development in Galaxy / Galaxy Interactive Tools
- Development in Galaxy / ToolFactory: Generating Tools From Simple Scripts
- Development in Galaxy / Scripting Galaxy using the API and BioBlend 📝
- Development in Galaxy / Data source integration
- Variant Analysis / Trio Analysis using Synthetic Datasets from RD-Connect GPAP
- Transcriptomics / Trajectory Analysis using Monocle3 📝
- Transcriptomics / Filter, Plot and Explore Single-cell RNA-seq Data 📝
- Transcriptomics / Generating a single cell matrix using Alevin 📝
- Transcriptomics / Inferring Trajectories using Python (Jupyter Notebook) in Galaxy 📝
- Transcriptomics / Combining datasets after pre-processing 📝
- Assembly / Genome Assembly of MRSA using Oxford Nanopore MinION Data 📝
- Assembly / De Bruijn Graph Assembly
- Assembly / Making sense of a newly assembled genome
- Assembly / Genome Assembly of MRSA using Illumina MiSeq Data 📝
- Sequence analysis / Mapping
- Teaching and Hosting Galaxy training / Galaxy Admin Training
- Teaching and Hosting Galaxy training / Training Infrastructure as a Service
- Teaching and Hosting Galaxy training / Running a workshop as instructor
- Teaching and Hosting Galaxy training / Organizing a workshop
- Contributing to the Galaxy Training Material / Creating a new tutorial
- Contributing to the Galaxy Training Material / Teaching Python ✍️
- Contributing to the Galaxy Training Material / Adding auto-generated video to your slides
- Contributing to the Galaxy Training Material / Updating diffs in admin training
- Contributing to the Galaxy Training Material / Tools, Data, and Workflows for tutorials
- Contributing to the Galaxy Training Material / GTN Metadata
- Contributing to the Galaxy Training Material / Including a new topic 📝
- Contributing to the Galaxy Training Material / Creating content in Markdown ✍️
- Contributing to the Galaxy Training Material / Adding Quizzes to your Tutorial
- Introduction to Galaxy Analyses / From peaks to genes
- Introduction to Galaxy Analyses / Galaxy 101
- Introduction to Galaxy Analyses / Data Manipulation Olympics 📝
- Galaxy Server administration / Galaxy Monitoring with gxadmin
- Galaxy Server administration / Use Singularity containers for running Galaxy jobs
- Galaxy Server administration / Galaxy Monitoring with Telegraf and Grafana
- Galaxy Server administration / Mapping Jobs to Destinations
- Galaxy Server administration / Distributed Object Storage
- Galaxy Server administration / Performant Uploads with TUS
- Galaxy Server administration / Training Infrastructure as a Service (TIaaS)
- Galaxy Server administration / External Authentication
- Galaxy Server administration / Galaxy Interactive Tools
- Galaxy Server administration / Galaxy Monitoring with Reports
- Galaxy Server administration / Ansible
- Galaxy Server administration / How I learned to stop worrying and love the systemd ✍️
- Galaxy Server administration / Reference Data with CVMFS
- Galaxy Server administration / Deploying a compute cluster in OpenStack via Terraform
- Galaxy Server administration / Reference Data with CVMFS without Ansible
- Galaxy Server administration / Deploying Tailscale/Headscale for private mesh networking ✍️
- Galaxy Server administration / Automation with Jenkins
- Galaxy Server administration / Connecting Galaxy to a compute cluster
- Galaxy Server administration / Deploying Wireguard for private mesh networking ✍️
- Galaxy Server administration / Running Jobs on Remote Resources with Pulsar
- Galaxy Server administration / Data Libraries
- Galaxy Server administration / Galaxy Installation with Ansible ✍️
- Galaxy Server administration / Galaxy Tool Management with Ephemeris
- Visualisation / Visualisation with Circos
- Visualisation / Genomic Data Visualisation with JBrowse
- Foundations of Data Science / Basics of using Git from the Command Line ✍️
- Foundations of Data Science / Python - Multiprocessing ✍️
- Foundations of Data Science / Python - Loops
- Foundations of Data Science / Advanced SQL
- Foundations of Data Science / CLI Educational Game - Bashcrawl
- Foundations of Data Science / Python - Math
- Foundations of Data Science / Virtual Environments For Software Development ✍️
- Foundations of Data Science / Python - Globbing
- Foundations of Data Science / Python - Argparse ✍️
- Foundations of Data Science / SQL with R
- Foundations of Data Science / CLI basics
- Foundations of Data Science / Python - Type annotations ✍️
- Foundations of Data Science / Python - Subprocess
- Foundations of Data Science / Python - Functions
- Foundations of Data Science / Advanced CLI in Galaxy
- Foundations of Data Science / dplyr & tidyverse for data processing
- Foundations of Data Science / SQL Educational Game - Murder Mystery
- Foundations of Data Science / Python - Lists & Strings & Dictionaries
- Foundations of Data Science / Python - Testing ✍️
- Foundations of Data Science / Variant Calling Workflow
- Foundations of Data Science / Conda Environments For Software Development ✍️
- Foundations of Data Science / Python - Introductory Graduation
- Foundations of Data Science / Python - Basic Types & Type Conversion
- Foundations of Data Science / Make & Snakemake
- Foundations of Data Science / Python - Files & CSV
- Foundations of Data Science / Introduction to SQL
- Foundations of Data Science / Python - Flow Control
- Foundations of Data Science / Python - Try & Except
- Foundations of Data Science / SQL with Python
- Foundations of Data Science / Version Control with Git 📝
- Using Galaxy and Managing your Data / Rule Based Uploader: Advanced
- Using Galaxy and Managing your Data / Name tags for following complex histories
- Using Galaxy and Managing your Data / Understanding Galaxy history system
- Using Galaxy and Managing your Data / Workflow Reports ⚙️
- Using Galaxy and Managing your Data / Rule Based Uploader
- Using Galaxy and Managing your Data / JupyterLab in Galaxy 📝
- Using Galaxy and Managing your Data / Using Workflow Parameters
- Using Galaxy and Managing your Data / Downloading and Deleting Data in Galaxy
- Using Galaxy and Managing your Data / Searching Your History
Slides
- Genome Annotation / Introduction to Genome Annotation
- Introduction to Galaxy Analyses / Introduction to Galaxy
- Galaxy Server administration / Galaxy from an administrator's point of view
- Visualisation / Visualisations in Galaxy
- Genome Annotation / Refining Genome Annotations with Apollo
- Development in Galaxy / Galaxy Interactive Environments
- Teaching and Hosting Galaxy training / Overview of the Galaxy Training Material for Instructors ✍️
- Contributing to the Galaxy Training Material / Creating Slides
- Introduction to Galaxy Analyses / A Short Introduction to Galaxy
- Galaxy Server administration / Galaxy Monitoring with gxadmin
- Galaxy Server administration / Galaxy Monitoring with Telegraf and Grafana
- Galaxy Server administration / Storage Management
- Galaxy Server administration / Ansible
- Galaxy Server administration / Server: Other
- Galaxy Server administration / Galaxy and Celery 📝
- Galaxy Server administration / Advanced customisation of a Galaxy instance
- Galaxy Server administration / Controlling Galaxy with systemd or Supervisor
- Galaxy Server administration / Galaxy Monitoring
- Galaxy Server administration / Reference Data with CVMFS
- Galaxy Server administration / Terraform
- Galaxy Server administration / Galactic Database
- Galaxy Server administration / Connecting Galaxy to a compute cluster
- Galaxy Server administration / Server Maintenance and Backups
- Galaxy Server administration / Empathy
- Galaxy Server administration / Running Jobs on Remote Resources with Pulsar
- Galaxy Server administration / User, Role, Group and Quota managment
- Galaxy Server administration / Storage Management
- Galaxy Server administration / Galaxy Tool Management with Ephemeris
- Visualisation / Circos
- Visualisation / JBrowse
- Introduction to Galaxy Analyses / Una breve introducción a Galaxy
- Introduction to Galaxy Analyses / Una Breve Introducción a Galaxy
News
Within the GTN we have had schemas for the various metadata we keep for a long time. It helps us automatically check community contributions to ensure they conform to all of the GTN standards, and will work correctly in our environment.
Full Story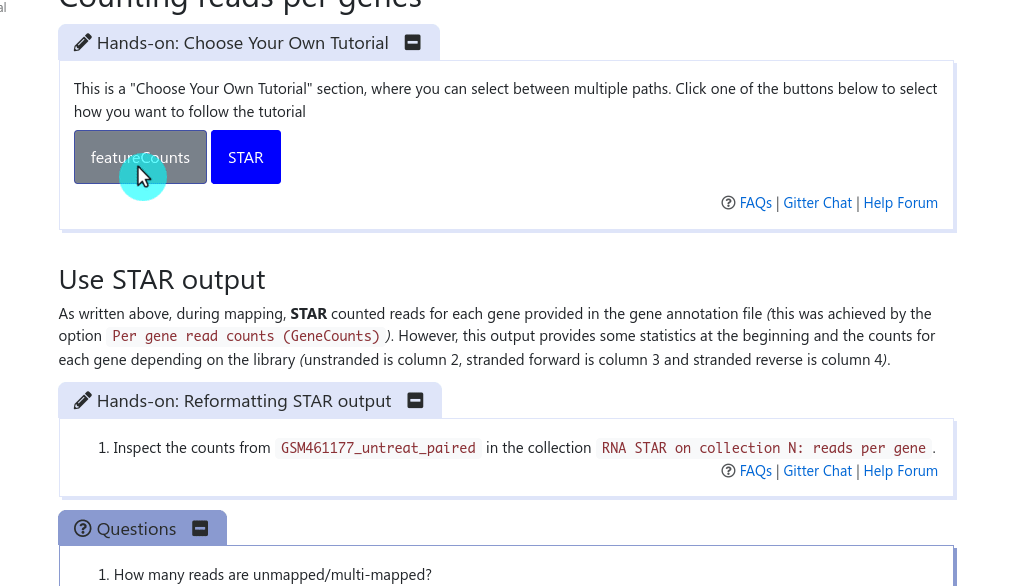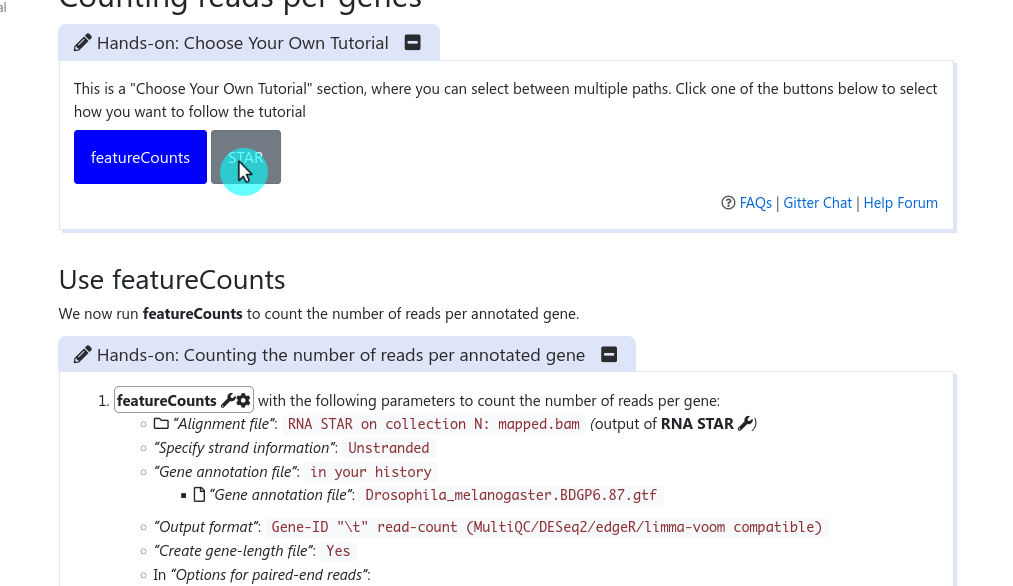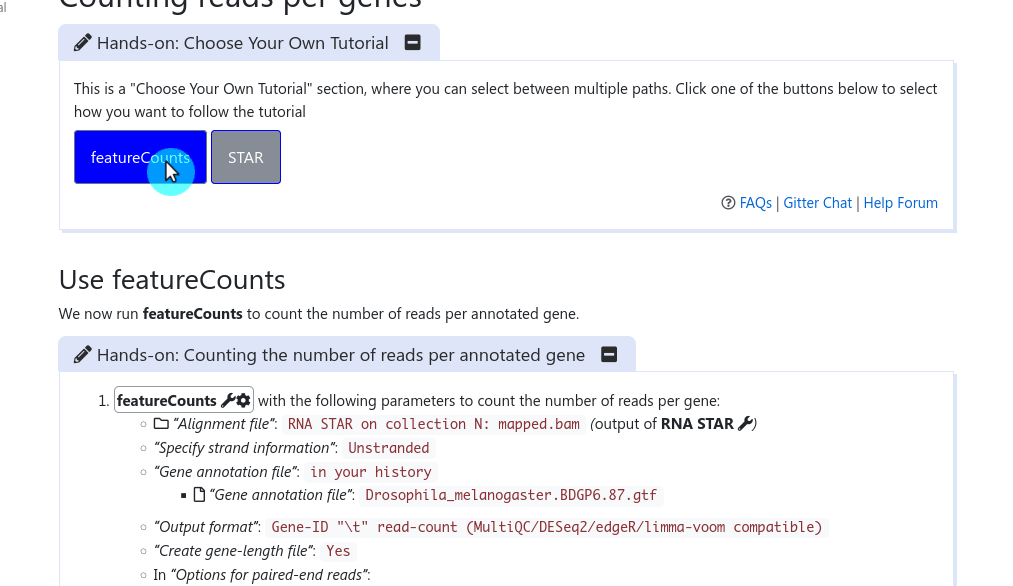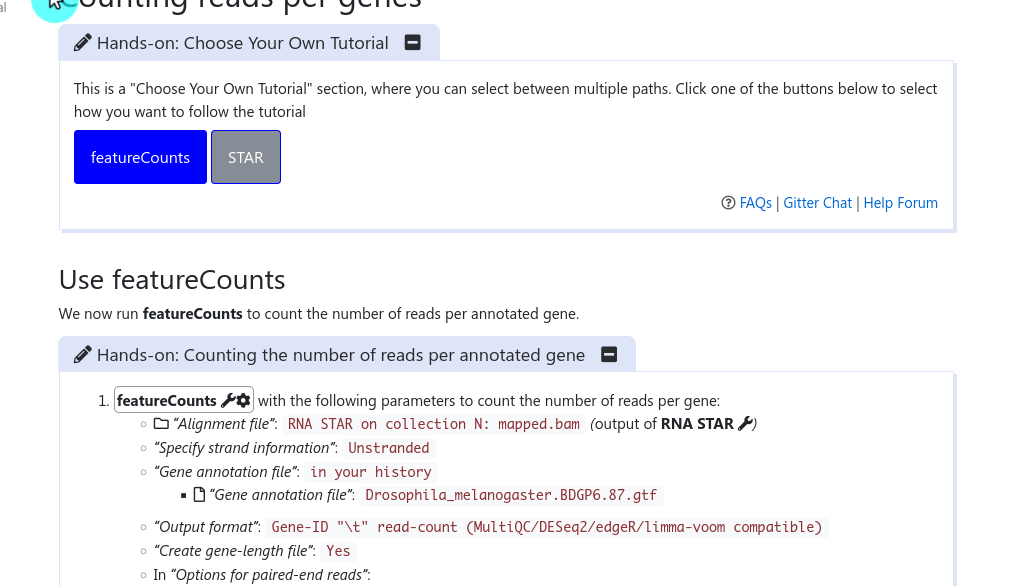
It has been a long discussed feature within the GTN Tutorial Author Community: how can we provide trainees with choice in their tutorials, and how can we as trainers not duplicate large tutorials just to show a slightly different path at one point?
Full Story
| Author(s) |
 |
Further building on the work in the automatic Jupyter Notebook, we’ve now re-written the Jupyter export to be faster, and more importantly added support for R and RMarkdown! Check out an example material. Here we take the content of the tutorial, written like normal GTN markdown, and we automatically convert it to Jupyter Notebooks and now RMarkdown documents! Check out the documentation on how to setup your tutorials to support this.
Full Story
| Author(s) |
 |
The Galaxy Training Network will now support annotating Funding Agencies on training materials. This is part of our ongoing effort to better annotate how people are contributing to the GTN and recognise all of the sources of contribution.
Full Story
The GTN does a lot of work to adhere to the best practices for FAIR training materials as set out in Garcia et al. 2020 and a lot of this work is behind-the-scenes or automatic from the infrastructure we provide, and we wanted to highlight that for educators and organisers who want to use GTN materials. This is one of the ways the GTN works to makes itself a fantastic platform training. Find out how contributing simple markdown tutorials can result in fantastically FAIR training.
Full Story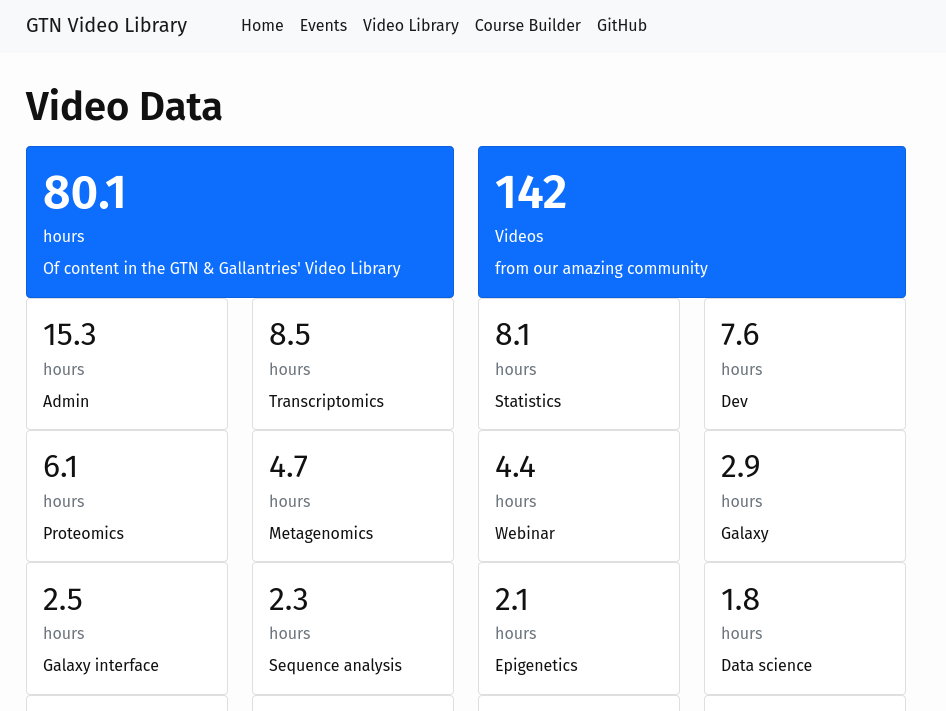
After runnning a number of virtual training events this year (e.g. Smörgåsbord, GCC2021), we now have a large collection of training videos around the GTN materials, created by this community.
Full Story
As part of the work for the Gallantries Grant, we wanted to access some of the data from the GTN in a more accessible, API based way. We are working on building a Video Library based on the wild success of our Smörgåsbord and GCC2021 events and to support that we wanted to start pulling in data from the GTN to save course authors time and effort when designing their asynchronous courses.
Full Story
Are you one of our amazing GTN Contributors? Were you ever bothered by how slow it was to build the site locally? This is now fixed completely! We have replaced one of the slowest steps of the GTN’s build process and rewritten it ourselves to be more performant.
Full Story
As part of our work under the Gallantries Grant we are establishing a new topic within the GTN specifically focused on Data Science skills. This topic will include tutorials covering things such as:
Full Story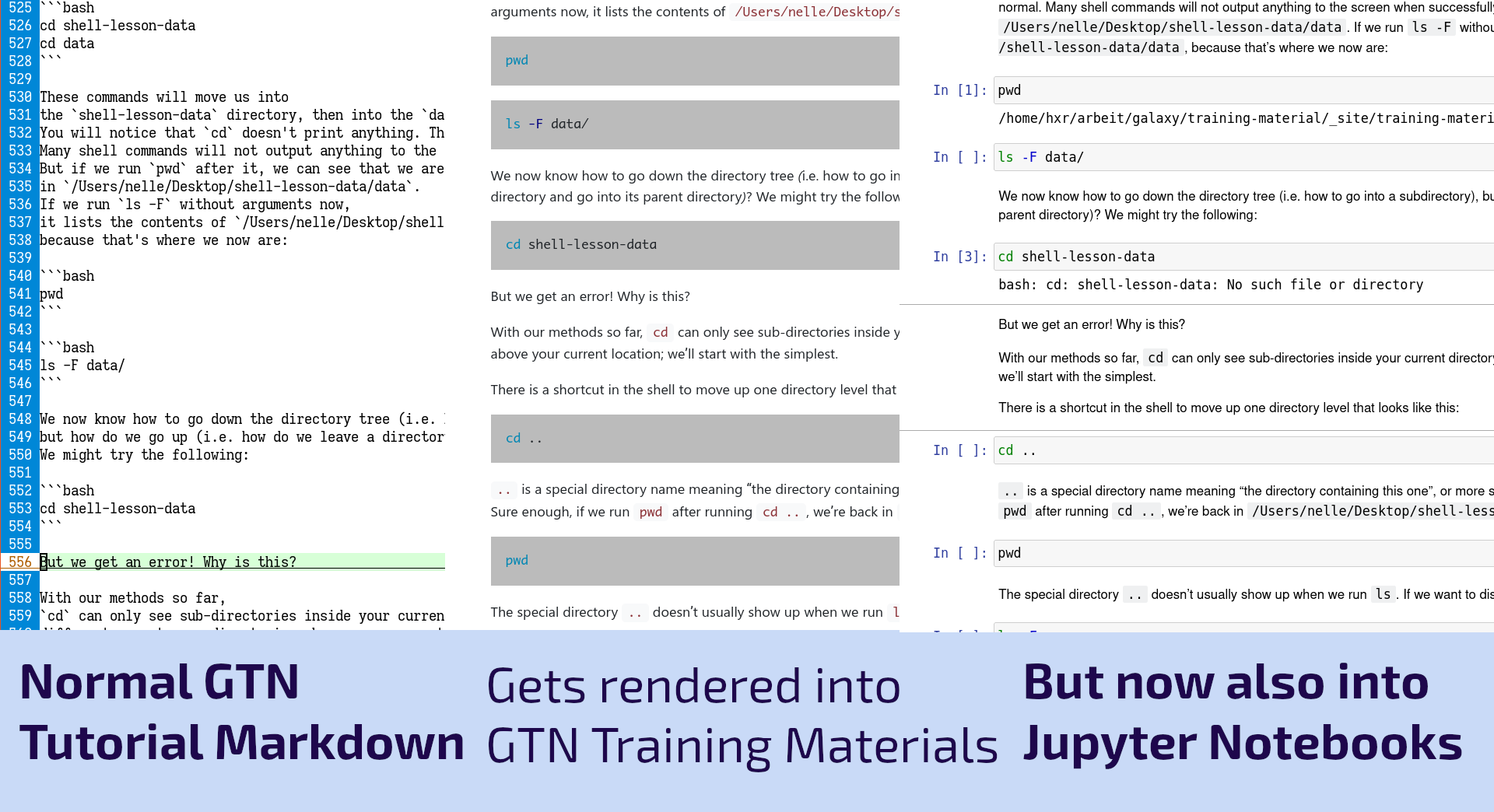
As part of the work for the Gallantries Grant, we found ourselves wanting to teach a number of non-Galaxy tutorials on topics such as Python, Bash, SQL, and Git. Writing the tutorials was always a bit awkward as you would spend a lot of time writing code blocks that would be copied and pasted by students into a terminal. We thought we could do better than that, so we decided we would try to convert these to Jupyter Notebooks where the students could read the training materials and simultaneously execute the code cells directly there with the text! This worked fantastically and now we’ve built this feature into the GTN to make it easier to run all of the coding tutorials.
Full Story
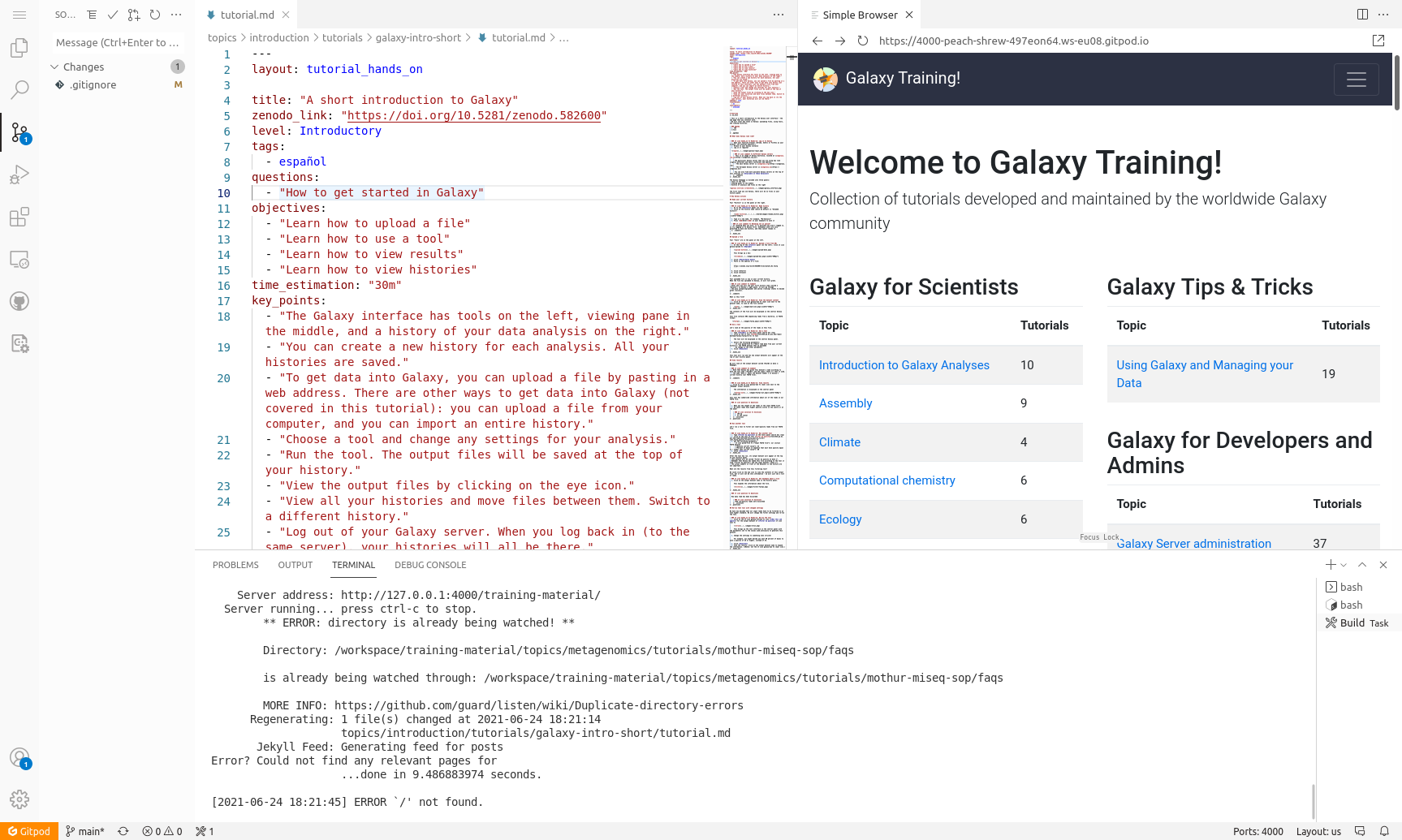
Want to contribute to the GTN without installing anything on your own computer? GitPod provides an online workspace for editing the GTN tutorials, and showing a live preview of the GTN website with your changes. This tutorial shows you how to set up GitPod, how to use it to get a live preview of the GTN website, and how to save any changes you make back to your GitHub fork. This makes it easier than ever to contribute to the GTN!
Full Story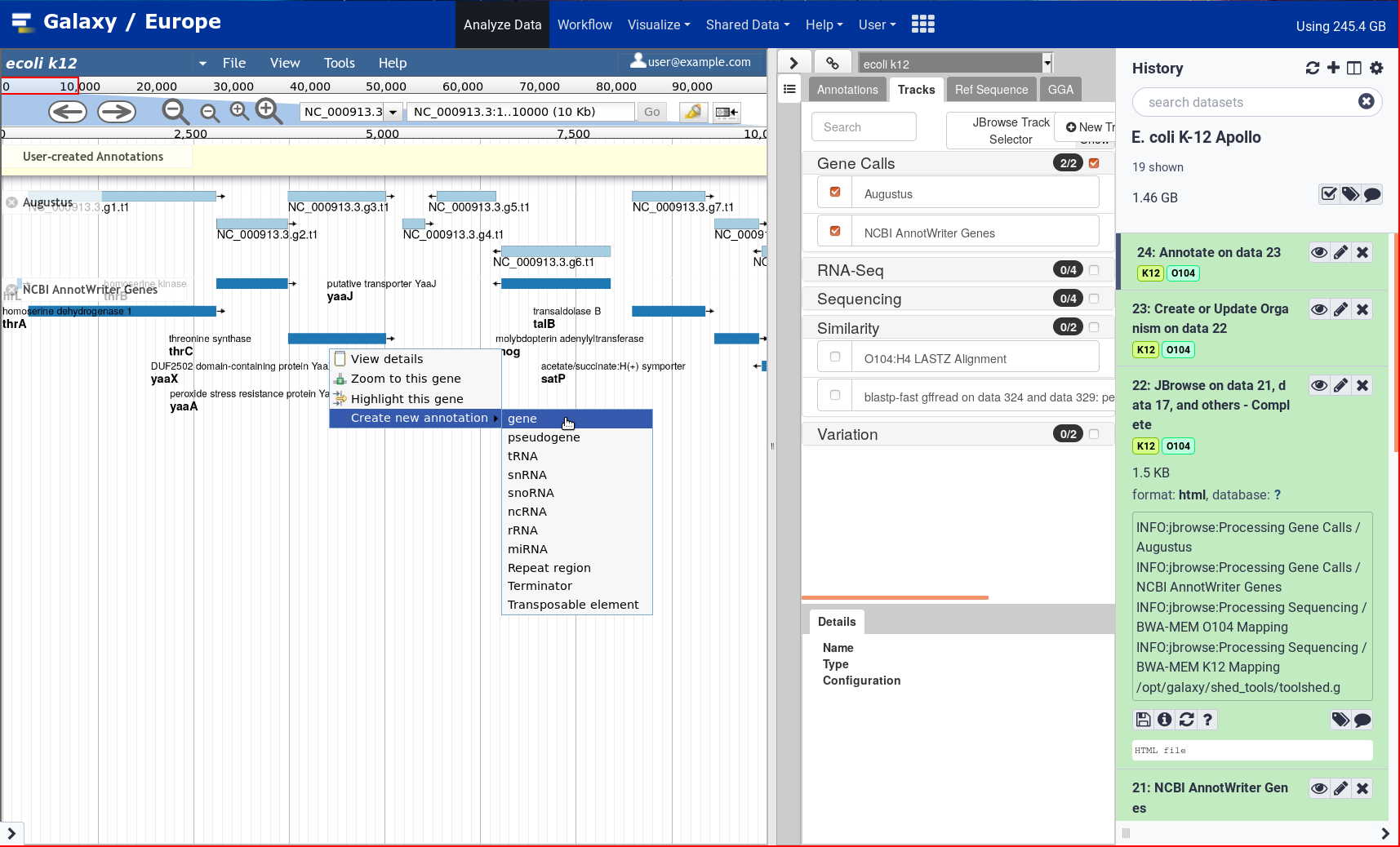
After two years, the Galaxy Genome Annotation team has finally finished their much awaited tutorial on doing Genome Annotation in Galaxy! Genome annotation is a time consuming and largely manual process as it requires the synthesis of a significant amount of varied information to make inferences, a process tha cannot be accomplished without an interactive editor. To support this, Apollo was deployed on UseGalaxy.eu. Over the past two years the GGA project has pushed forward apollo and tooling around it, and it finally culminates in this look at how to annotated your genomes in Apollo.
Full Story
Did you ever start teaching a GTN tutorial, only to realise something changed recently? Oh no! A while ago the GTN created an Archive to address just this concern. On every tag, we’d build a copy of the website which we would place online and never touch again. This way we’d have permanent online copies so that if someone needed to go back to an older version of the GTN, they could.
Full Story
Thanks to the great tutorial developed by first time contributor Erik Schill and edited by Simon Bray , we noticed that they defined a number of abbreviations and re-used those throughout their tutorial.
Full Story
¡El primer tutorial en español ya está disponible! Galaxy siempre ha tenido tutoriales traducidos automáticamente mediante Google-Translate, ahora nos estamos embarcando en un nuevo proyecto para estudiar la experiencia de aprendizaje con tutoriales en bioinformática traducidos por humanos. Nuestro objetivo es crear un paquete de tutoriales para el análisis de datos single-cell (célula única) en primera instancia, con el fin de estudiar su uso y utilidad en un taller para hablantes nativos de español. Agradecemos enormemente a las nuevas participantes Patricia Carvajal López y Alejandra Escobar-Zepeda (EMBL-EBI) así como a las miembros existentes de GTN Beatriz Serrano-Solano (University of Freiburg), Saskia Hiltemann & Helena Rasche (Erasmus-MC), por su arduo trabajo en este proyecto de traducción y mantenimiento y, por supuesto, a Wendi Bacon (The Open University/EBI) como líder de proyecto.
Full Story
One of our contributors ( Mehmet Tekman ) found himself wanting to link to a specific time point in a video. We said “Wow, what a fantastic idea!” and then we implemented it. To support that we developed a very simple standalone video player page where you can view the GTN videos and browse through our catalog. It features the full transcript on the left, some suggested videos on the right, and a very standard <video> player.
Full Story
Two new assembly tutorials have been added to the GTN! These tutorials walk you through the process of assembling MRSA strains from real-world datasets. MRSA stands for “Methicillin- resistant Staphylococcus aureus, it is resistance to several antibiotics and is a common source of hospital-acquired infections and outbreaks. In this tutorial you will assess the quality of your data, perform an assembly and assess its quality, analyze your data for the presence of antimicrobial resistance (AMR) genes, and annotate the genome using Prokka and JBrowse.
Full Story
Snippets have received an major overhaul! Snippets/FAQs are small reusable bits of training, often answering a single question. In addition to the general Galaxy snippets, we now also support topic-level and tutorial-level FAQs. This will allow contributors to add common questions and answers to their tutorials, which not only can be included in the tutorial text itself, but are also used to autogenerate an FAQ page for the tutorial or topic listing all common questions (and answers).This will be useful both to participants following tutorials via self-study, and also to instructors preparing for a training event.
Full Story
Every 3 months we organise one day dedicated to the GTN community! Thursday, May 20th is the next GTN CoFest. We will be working on the training materials, have discussions with the global Galaxy traning community. Do you teach with Galaxy? Want to learn how to add your tutorial to the GTN? New to the community and just want to learn more? Everybody is welcome!
Full Story
The GTN instructors community has just run the largest-ever Galaxy training event: “the GTN Smörgåsbord: A Global Galaxy Course”, co-organized with the Gallantries and the CINECA project and the Galaxy Training Network.
Full Story
This past year has been a struggle for all of us, but one task has become apparent as one of the most time consuming portions of course preparation during a pandemic: Video Production.
Full Story