name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/visualisation" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a class="nav-link" href="https://github.com/galaxyproject/training-material/edit/main/topics/visualisation/tutorials/circos/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTN.png" alt="Galaxy Training Network" class="cover-logo"/> # Circos <div markdown="0"> <div class="contributors-line"> Authors: <a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-hexylena"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/hexylena?s=27" alt="Avatar">Helena Rasche</a> <a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-shiltemann"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/shiltemann?s=27" alt="Avatar">Saskia Hiltemann</a> <a href="/training-material/hall-of-fame/erasmusplus/" class="contributor-badge contributor-erasmusplus"><img src="https://avatars.githubusercontent.com/erasmusplus?s=27" alt="Avatar">Erasmus+ Programme</a> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"><i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: Dec 14, 2021</div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 6em;"> <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ## Requirements Before diving into this slide deck, we recommend you to have a look at: - [Introduction to Galaxy Analyses](/training-material/topics/introduction) --- ### <i class="far fa-question-circle" aria-hidden="true"></i><span class="visually-hidden">question</span> Questions - What is Circos? - How can I use Circos within Galaxy? - What are some of the different track types I can plot? --- # Circos for Genomics Visualisation --- ### What is Circos? - Circular Graphics - Highly flexible, but also very complex configuration - ... but beautiful visualisations - Can visualise a huge range of data types 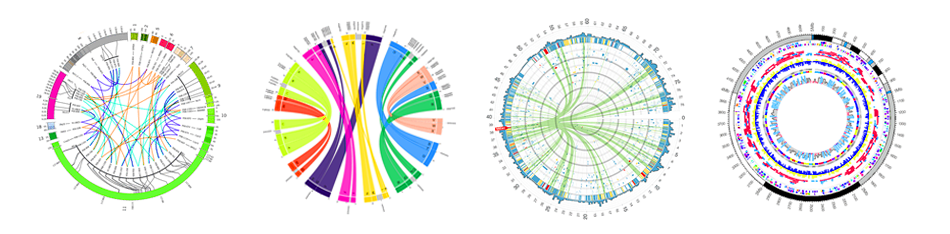 ??? Many researchers find Circos overwhelming and complex. This tool can save them time and we will hopefully show them how to reproduce several plots from published papers. --- ### Circos in Galaxy - Tool that produces a Circos Plot (PNG / SVG) - Circos is too complex to put all configuration options in Galaxy - Option to **download full configuration** directory - allows for further manual tweaking locally - 90% done inside Galaxy, last 10% finished locally .image-50[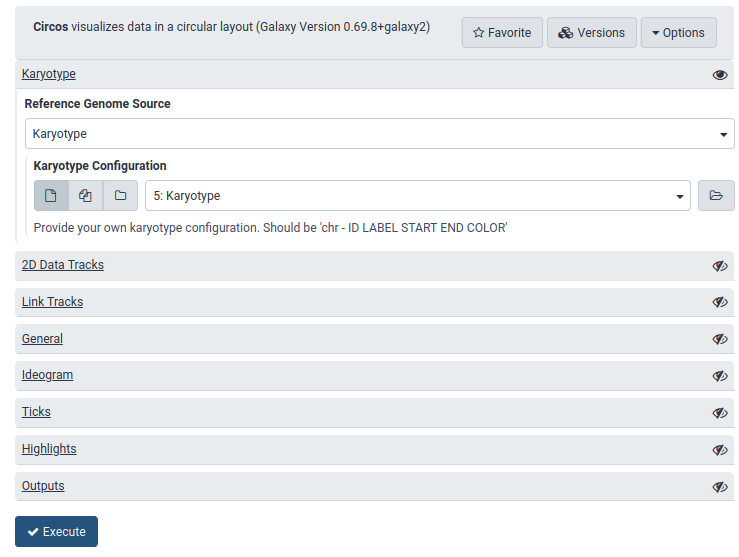] ??? Emphasize that producing production quality plots is still a lot of work. Not all configuration options available in Circos can be set in the Galaxy tool. But all the config files can be downloaded so that you can tweak the plot manually if you have circos installed on your computer We cannot support them uploading these edited config files into Galaxy because of potential security issues (circos can shell out.) --- ### Supported Features - Covers *most* of the possibilities of Circos - Can **auto convert** GFF3/BED/(big)Wig datasets into appropriate formats - Supports common Circos features like: - histograms - heatmaps - "tile" (from gff3/bed) - line - scatter - ribbons - Complex Circos visualisations made easy (Thanks Galaxy!) --- ### Tracks: Histograms - For data which associate a position with a value - Can be good for - Gene density on large genomes - Variation density? - GC Skew 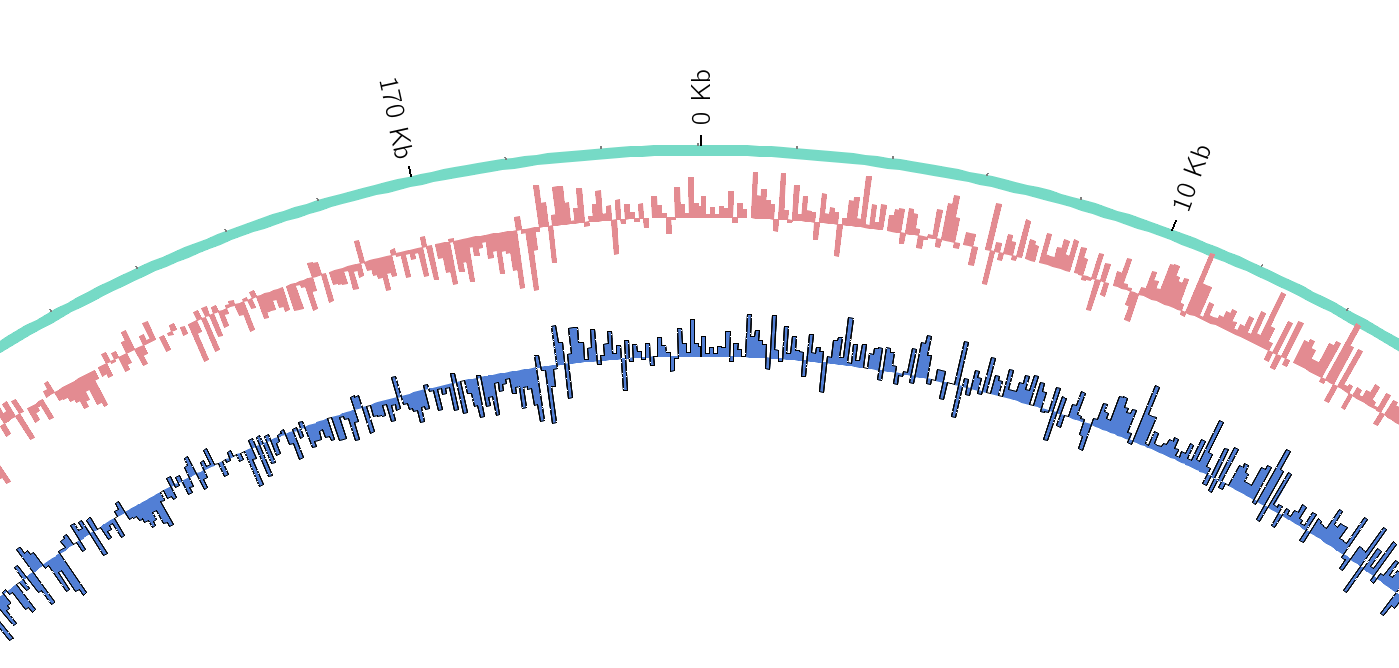 ??? --- ### Tracks: Heatmaps - Similar use case to histograms - Histograms may provide easier quantification for viewers 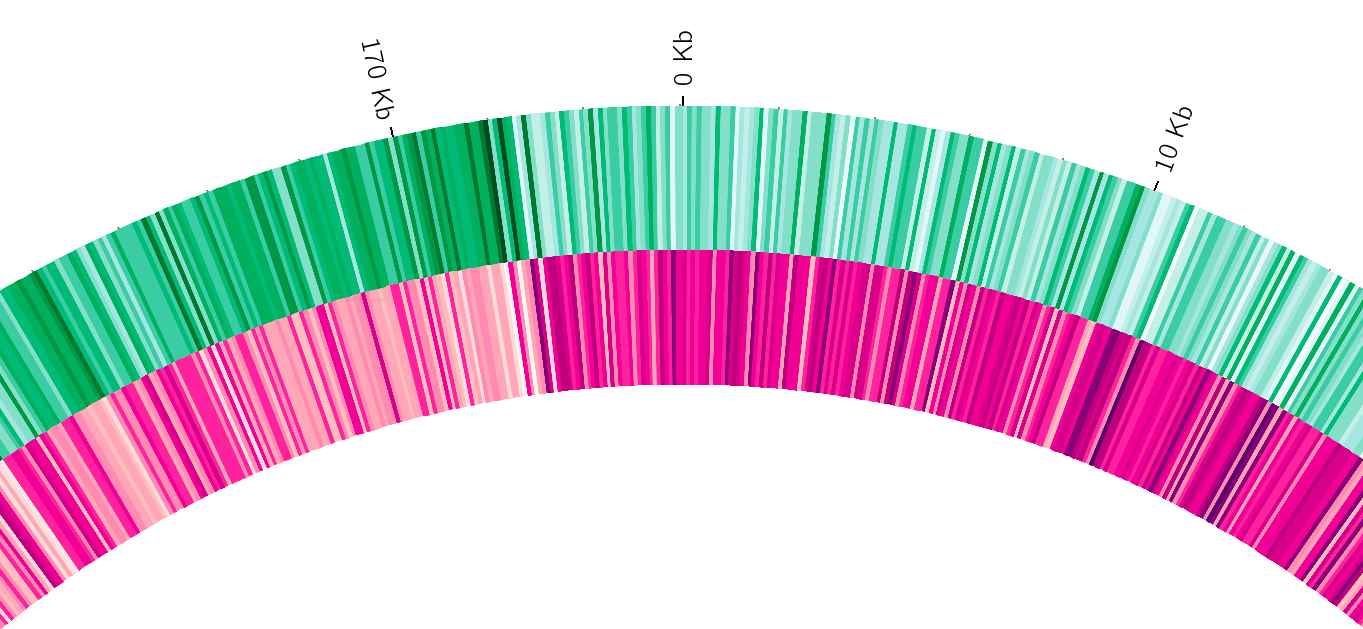 ??? It can be easier to quantify differences between two values next to each other with the histogram than relying on the viewer's ability to distinguish + quantify how far apart two colours lie on a colour scale. These can be pretty but are harder to extract data from --- ### Tracks: Tiles - Show genomic regions - Great for genes on small genomes 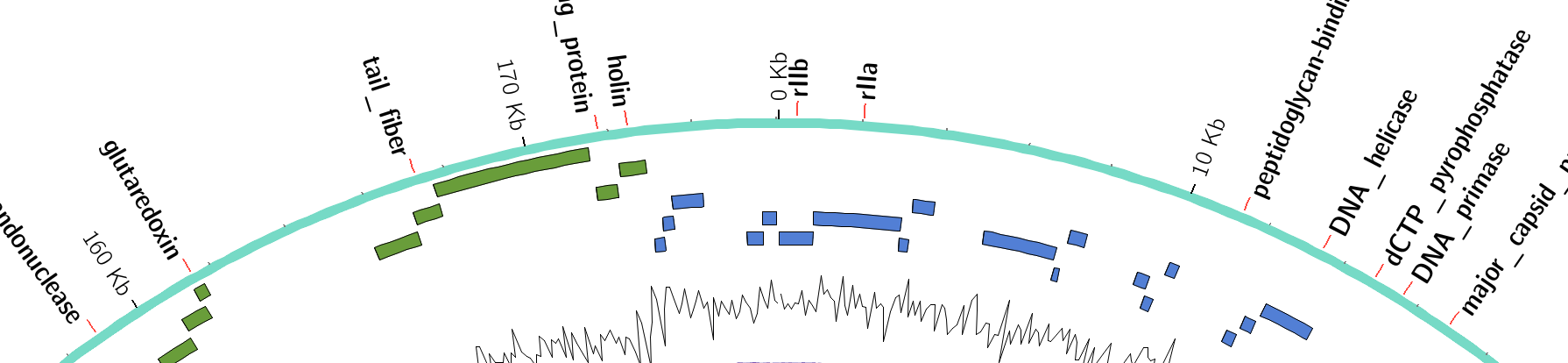 ??? "Tile tracks are used to show spans such as genomic regions (genes, exons, duplications) or coverage elements (clones, sequence reads). Tiles will stack within their track to avoid overlap." (circos documentation) --- ### Tracks: Scatter - Can be used similarly to heatmaps and histograms .") ??? Image is based off of the Circos Scatter plot lesson. All images in this tutorial should be reproducible with the Galaxy Circos tool http://circos.ca/documentation/tutorials/2d_tracks/scatter_plots/lesson --- ### Tracks: Lines 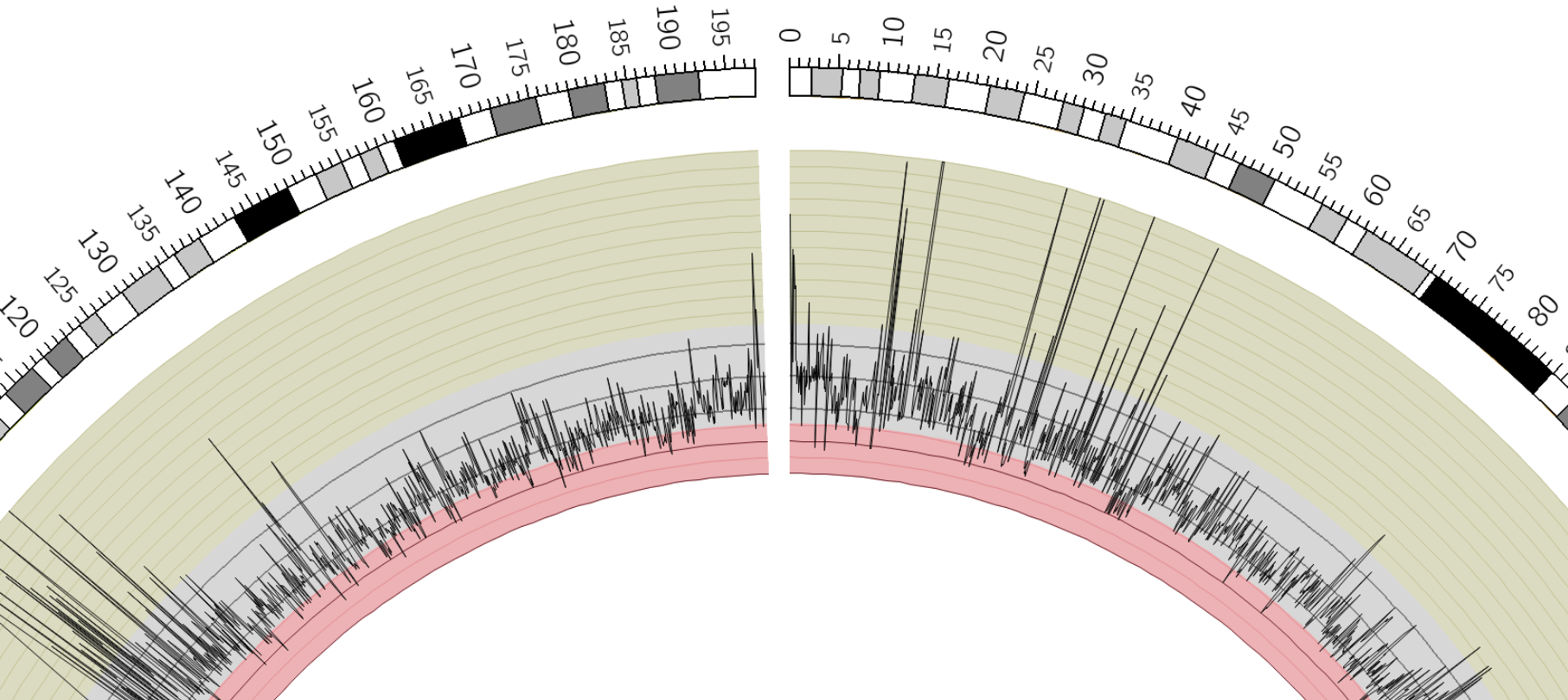 ??? Image is based off of the Circos Line lesson. http://circos.ca/documentation/tutorials/2d_tracks/line_plots/images --- ### Tracks: Ribbons - Shows a relationship between two regions - On the same chromosome or on different ones - Great for synteny mapping or other regional interactions 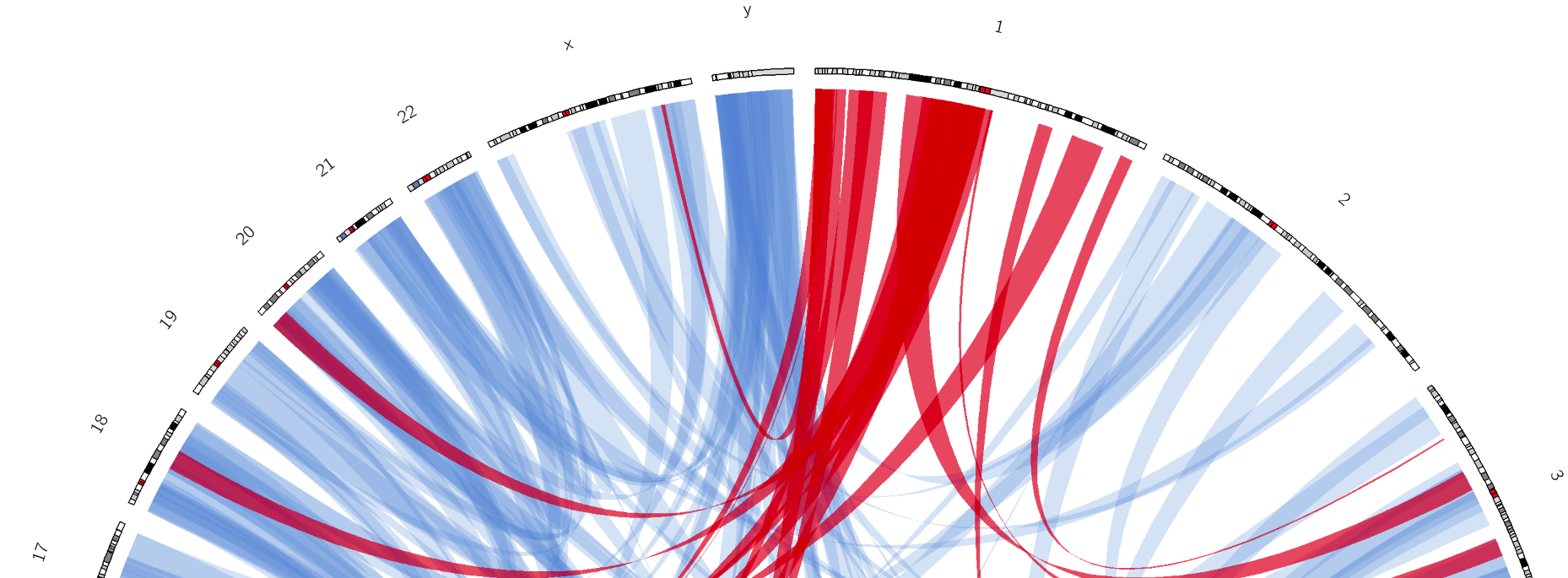 ??? This plot highlights all interactions coming from, or going to chr1. This plot was generated from a set of random links. The circos-tools package includes a `randomlinks` tool that will generate these files. There is no galaxy wrapper for this tool. --- ### Circos: An iterative process! - Because of its complexity, creating a Circos plot is often an *iterative* process - Build your plot one step at a time, check the output regularly! .image-40[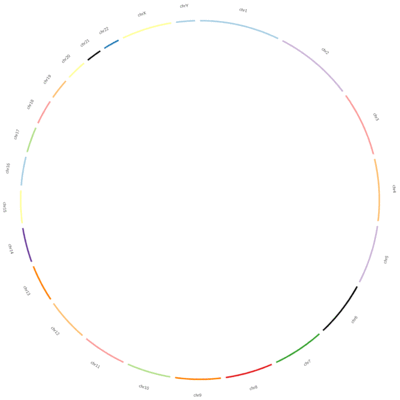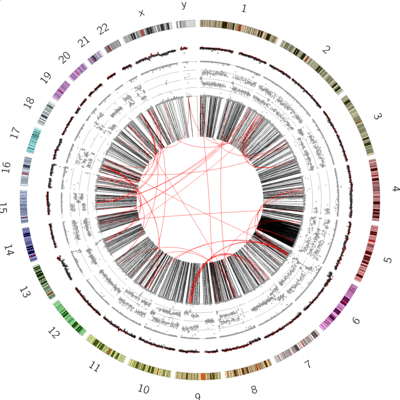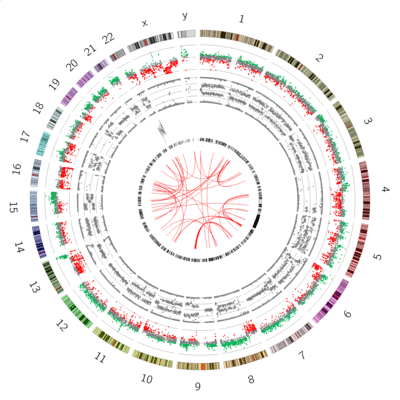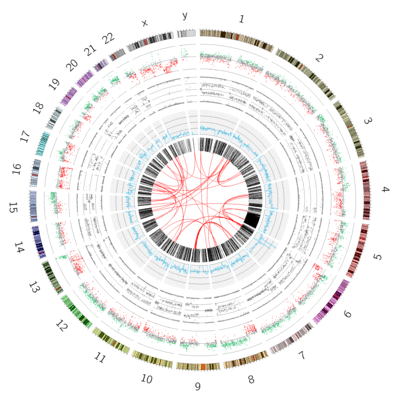] --- ### <i class="fas fa-key" aria-hidden="true"></i><span class="visually-hidden">keypoints</span> Key points - Circos is very powerful, but also very complex - Creating plots is an iterative process - The full configuration directory can be downloaded from Galaxy to be further tweaked locally --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> Authors: <a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-hexylena"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/hexylena?s=27" alt="Avatar">Helena Rasche</a> <a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-shiltemann"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/shiltemann?s=27" alt="Avatar">Saskia Hiltemann</a> <a href="/training-material/hall-of-fame/erasmusplus/" class="contributor-badge contributor-erasmusplus"><img src="https://avatars.githubusercontent.com/erasmusplus?s=27" alt="Avatar">Erasmus+ Programme</a> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTN.png" alt="Galaxy Training Network" style="height: 100px;"/> </div> <a rel="license" href="https://creativecommons.org/licenses/by/4.0/"> This material is licensed under the Creative Commons Attribution 4.0 International License</a>.