Data source integration
OverviewQuestions:Objectives:
How can I write a tool that can import data into Galaxy from an external database?
What are “data sources” and how do they function?
Is there any ready-to-use example?
Time estimation: 10 minutesSupporting Materials:Last modification: Sep 28, 2022
Data Source Integration
An important goal of Galaxy is scalability. A major bottleneck when it comes to analysis of big data sets is the time and space it takes of copying these data sets.
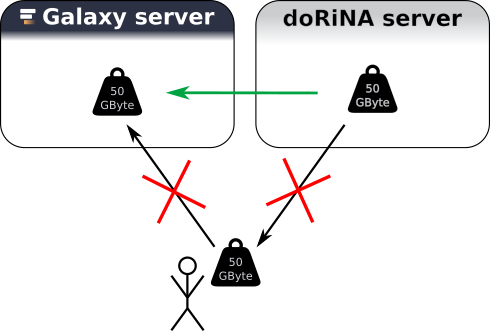
Galaxy provides an interface such that it can communicate with other servers to get data directly into the Galaxy environment of a user without the need of “downloading” the data. In this hands on, we will use the resource from DoRiNA Server (Blin et al. 2014). but the main point about this short section is: if you have a data source which you think is very important for your research with Galaxy let us know!
Hands-on: Hands on!
- Create a new history called “doRiNA”
- Go to Get Data::doRiNA search
- Choose hg19 from the drop-down list -> Search Database
- Leave everything as is and choose from the Regulators (set A) drop-down list “hsa-let-7astar-CLASH” -> Search doRiNA
- Use the “Send to Galaxy” button
- Notice the new History Item
You can see the tutorial section of the DoRiNA website for more detailed examples. That was very easy for all of you! If you want your database of choice to be accessible as easy as this let us know!
Key points
It is possible to couple an external data resource with a Galaxy server
The external data resource is accessed through his native interface
Data flows from the external data resource to the Galaxy server without the need of “downloading” the data
Frequently Asked Questions
Have questions about this tutorial? Check out the tutorial FAQ page or the FAQ page for the Development in Galaxy topic to see if your question is listed there. If not, please ask your question on the GTN Gitter Channel or the Galaxy Help ForumReferences
- Blin, K., C. Dieterich, R. Wurmus, N. Rajewsky, M. Landthaler et al., 2014 DoRiNA 2.0—upgrading the doRiNA database of RNA interactions in post-transcriptional regulation. Nucleic Acids Research 43: D160–D167. 10.1093/nar/gku1180
Feedback
Did you use this material as an instructor? Feel free to give us feedback on how it went.
Did you use this material as a learner or student? Click the form below to leave feedback.
Citing this Tutorial
- Bérénice Batut, Saskia Hiltemann, Gianmauro Cuccuru, Helena Rasche, 2022 Data source integration (Galaxy Training Materials). https://training.galaxyproject.org/training-material/topics/dev/tutorials/data-source-integration/tutorial.html Online; accessed TODAY
- Batut et al., 2018 Community-Driven Data Analysis Training for Biology Cell Systems 10.1016/j.cels.2018.05.012
Congratulations on successfully completing this tutorial!@misc{dev-data-source-integration, author = "Bérénice Batut and Saskia Hiltemann and Gianmauro Cuccuru and Helena Rasche", title = "Data source integration (Galaxy Training Materials)", year = "2022", month = "09", day = "28" url = "\url{https://training.galaxyproject.org/training-material/topics/dev/tutorials/data-source-integration/tutorial.html}", note = "[Online; accessed TODAY]" } @article{Batut_2018, doi = {10.1016/j.cels.2018.05.012}, url = {https://doi.org/10.1016%2Fj.cels.2018.05.012}, year = 2018, month = {jun}, publisher = {Elsevier {BV}}, volume = {6}, number = {6}, pages = {752--758.e1}, author = {B{\'{e}}r{\'{e}}nice Batut and Saskia Hiltemann and Andrea Bagnacani and Dannon Baker and Vivek Bhardwaj and Clemens Blank and Anthony Bretaudeau and Loraine Brillet-Gu{\'{e}}guen and Martin {\v{C}}ech and John Chilton and Dave Clements and Olivia Doppelt-Azeroual and Anika Erxleben and Mallory Ann Freeberg and Simon Gladman and Youri Hoogstrate and Hans-Rudolf Hotz and Torsten Houwaart and Pratik Jagtap and Delphine Larivi{\`{e}}re and Gildas Le Corguill{\'{e}} and Thomas Manke and Fabien Mareuil and Fidel Ram{\'{\i}}rez and Devon Ryan and Florian Christoph Sigloch and Nicola Soranzo and Joachim Wolff and Pavankumar Videm and Markus Wolfien and Aisanjiang Wubuli and Dilmurat Yusuf and James Taylor and Rolf Backofen and Anton Nekrutenko and Björn Grüning}, title = {Community-Driven Data Analysis Training for Biology}, journal = {Cell Systems} }

 Questions:
Questions:
