Contributions
The following list includes only slides and tutorials where the individual has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
GitHub Activity
Tutorials
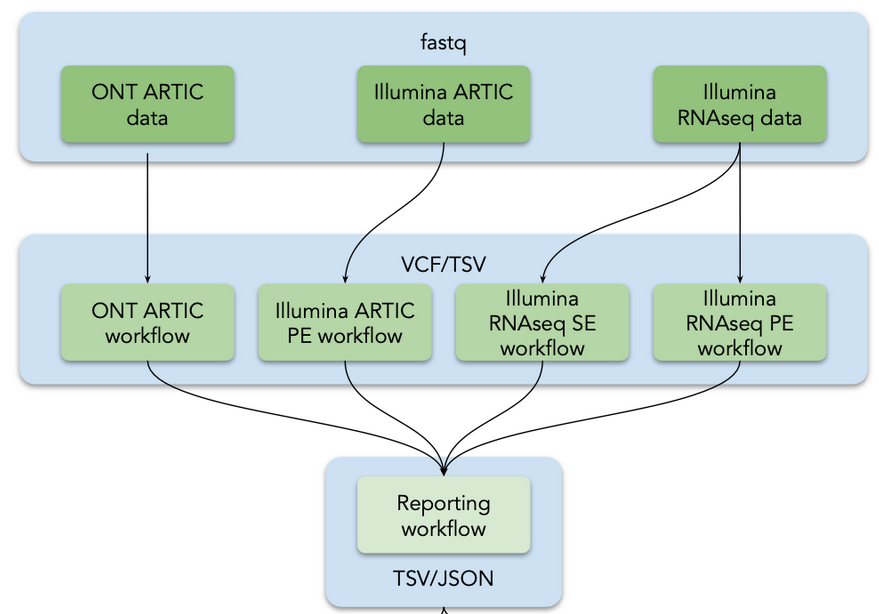
- Variant Analysis / Mutation calling, viral genome reconstruction and lineage/clade assignment from SARS-CoV-2 sequencing data
- Variant Analysis / Trio Analysis using Synthetic Datasets from RD-Connect GPAP
- Variant Analysis / Exome sequencing data analysis for diagnosing a genetic disease
- Variant Analysis / Identification of somatic and germline variants from tumor and normal sample pairs
- Variant Analysis / Somatic Variant Discovery from WES Data Using Control-FREEC
- Variant Analysis / Mapping and molecular identification of phenotype-causing mutations
- Transcriptomics / Pre-processing of Single-Cell RNA Data
- Assembly / Unicycler assembly of SARS-CoV-2 genome with preprocessing to remove human genome reads
- Sequence analysis / Removal of human reads from SARS-CoV-2 sequencing data
- Using Galaxy and Managing your Data / Automating Galaxy workflows using the command line
News
Authors:
Effectively monitoring global infectious disease crises, such as the COVID-19 pandemic, requires capacity to generate and analyze large volumes of sequencing data in near real time. These data have proven essential for monitoring the emergence and spread of new variants, and for understanding the evolutionary dynamics of the virus.
Full Story