Generating theoretical possible pathways for the production of Lycopene in E.Coli using Retrosynthesis tools
OverviewQuestions:Objectives:
Which heterologous pathways are candidates to produce a compound in a chassis?
Requirements:
Define what data are required to run RetroSynthesis analysis.
Understanding how to run the tools to search heterologous pathways.
Time estimation: 30 minutesSupporting Materials:Last modification: Oct 18, 2022
Introduction
Galaxy-SynBioCAD portal is the first toolshed for synthetic biology, metabolic engineering, and industrial biotechnology (Hérisson et al. 2022). It provides a set of Retrosynthesis tools aimed at finding pathways to synthesize heterologous compounds in chassis organisms (RetroRules (Duigou et al. 2018), RetroPath2.0 (Delépine et al. 2018), RP2Paths, rpCompletion).
Retrosynthesis is a concept originally proposed for synthetic chemistry where chemists have to work backwards, starting from a target product to reach precursors that are endogenous to the chassis (host organism).
Typically, the target compound, also named “source compound” is the compound of interest one wishes to produce, while the precursors are usually compounds that are natively present in a chassis strain.
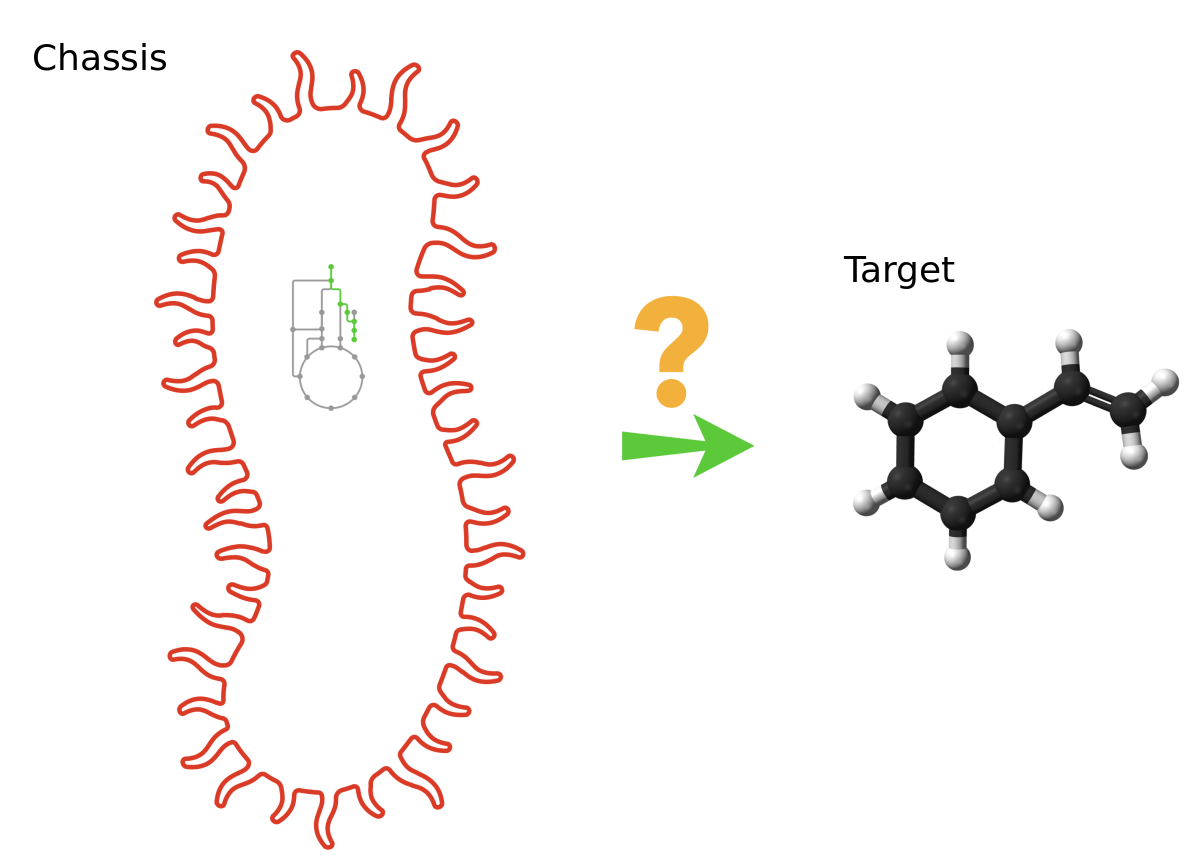
In this tutorial, we want to obtain the reactions producing the lycopene (source) into the iML1515 Escherichia Coli strain (chassis).
To do that, we will use the following RetroSynthetis Workflow composed of 3 key steps.
First, we aggregate the metabolites present in the chassis and download reaction rules.
Then, RetroPath2.0 generates feasible metabolic routes between a collection of chemical species contained within a GEM SBML (Systems Biology Markup Language) file of the selected organism, a target molecule that the user wishes to produce, and reaction rules extracted from RetroRules.
Lastly, the metabolic network is then deconstructed into individual pathways using RP2paths and rpCompletion takes those individual metabolic pathways to filter them (duplicated pathways are removed), splits them into sub-pathways by adding the appropriate cofactors, and finally converted them to SBML files.
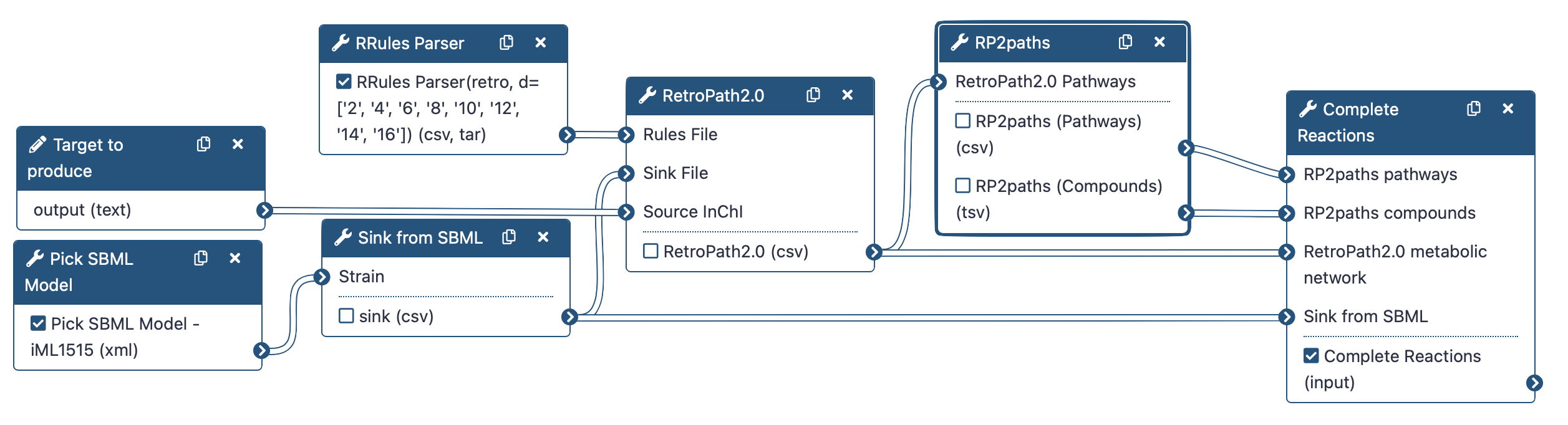
Note that we will run the steps of this workflow individually so as not to neglect the understanding of the intermediate steps as well. Then, we will run the workflow automatically so that it itself retrieves the outputs from the previous step and gives them as input to the next tool.
AgendaIn this tutorial, we will cover:
Data Preparation
RetroSynthesis workflow will be run with the following inputs:
- The International Chemical Identifier (InChI) of the compound of interest to produce,
- The structure of metabolites present in the chosen chassis (E. coli),
- Reaction rules (generated by RRules Parser node that calls RetroRules).
The data used are pretty straight forward to obtain.
Firstly, we download an SBML model, then we select all sinks to use into the RetroPath2.0 software from this model.
Lastly, we request from RetroRules all possible reactions to find a chemical reaction cascade that produces the target.
Download a model
Hands-on: Select a model.
- Run Pick SBML Model Tool: toolshed.g2.bx.psu.edu/repos/tduigou/get_sbml_model/get_sbml_model/0.0.1 with the following parameters:
- galaxy-dropdown “Strain”:
Escherichia coli str. K-12 substr. MG1655 (iML1515)Comment: What does this tool do?The selected SBML model is downloaded from the BiGG database.
Question
- What is the file format of the model?
- The SBML is based on XML.
Create a sink file from the SBML model
Hands-on: Generate a sink file.
- Run Sink from SBML Tool: toolshed.g2.bx.psu.edu/repos/tduigou/rpextractsink/rpextractsink/5.12.1 with the following parameters:
- param-file “Strain”:
sbml_model(output of Pick SBML Model tool)- “SBML compartment ID”:
cComment: Choose a compartment corresponding to your modelYou can specify the compartment from which the tool will extract the chemical species. The default is
c, the BiGG code for the cytoplasm. If the user wishes to upload an SBML file from another source, then this value must be changed.
Question
- What this tool does?
- How many columns are in the file?
- This tool creates a friendly CSV file format that can be used as sink input for RetroPath2.0.
- Click on galaxy-eye, you should see 2 columns: “Name” and “InChi”
Retrieve the reaction rules
Hands-on: Generate a file with all reactions.
- Run RRules Parser Tool: toolshed.g2.bx.psu.edu/repos/tduigou/rrparser/rrparser/2.4.6 with the following parameters:
- “Rule Type”:
retro- “Select the diameters of the reactions rules”:
2,4,6,8,10,12,14and16- galaxy-toggle “Compress output”:
noComment: How to choose a right diameter ?The diameter of the sphere including the atoms around the reacting center. The higher is the diameter, the more specific are the rules.
Question
- Does a low diameter select specific rules?
- How many rows are in the file?
- No, a low diameter selects more unspecific rules.
- More than 200 thousands!
Run the Retrosynthesis algorithm
RetroPath2.0 is an open-source tool for building retrosynthesis networks by combining reaction rules and a retrosynthesis-based algorithm to link the desired target compound to a set of available precursors. The RetroPath2.0 tool is freely available at myExperiment. The retrosynthesis network is outputted as a CSV file providing reactions in the reaction SMILES format and chemicals in both SMILES and InChI formats along with other information like the score for each reaction.
Launch RetroPath2.0
Hands-on: Build a reaction network
- Run RetroPath2.0 Tool: toolshed.g2.bx.psu.edu/repos/tduigou/retropath2/retropath2/2.3.0 with the following parameters:
- param-file “Rules File”:
out_rules(output of RRules Parser tool)- param-file “Sink File”:
sink(output of Sink from SBML tool)- “InChI type”:
By string
- “Source InChI”: InChI=1S/C40H56/c1-33(2)19-13-23-37(7)27-17-31-39(9)29-15-25-35(5)21-11-12-22-36(6)26-16-30-40(10)32-18-28-38(8)24-14-20-34(3)4/h11-12,15-22,25-32H,13-14,23-24H2,1-10H3/b12-11+,25-15+,26-16+,31-17+,32-18+,35-21+,36-22+,37-27+,38-28+,39-29+,40-30+
Be careful, you must have
InChi=in front of your InChi key.
Question
- What is the file format produce by RetroPath2.0?
- A csv file.
Enumerate pathways with RP2paths
The RetroPath2.0 algorithm produces a reaction network, we want to have one pathway per file in SBML format. We need to split the network and perform some adjustments to these pathways.
Split the network
Hands-on: Build a reaction network
- Run RP2paths Tool: toolshed.g2.bx.psu.edu/repos/tduigou/rp2paths/rp2paths/1.5.0 with the following parameters:
- param-file “RetroPath2.0 Pathways”:
Reaction_Network(output of RetroPath2.0 tool)Comment: PrincipleExtracts the set of pathways that lies in a metabolic space file output by the RetroPath2.0 workflow
Question
- Why producing multiple pathways?
- How many outputs are produced?
- One network could represent several pathways, so several solutions.
- Two outputs are produced: one corresponding to the metabolites, the other one corresponding to the pathways.
Refine reactions
Hands-on: Refine reactions
- Run Complete Reactions Tool: toolshed.g2.bx.psu.edu/repos/tduigou/rpcompletion/rpcompletion/5.12.2 with the following parameters:
- param-file “RP2paths pathways”:
master_pathways(output of RP2paths tool)- param-file “RP2paths compounds”:
compounds(output of RP2paths tool)- param-file “RetroPath2.0 metabolic network”:
Reaction_Network(output of RetroPath2.0 tool)- param-file “Sink from SBML”:
sink(output of Sink from SBML tool)Comment: PrincipleEach reaction rule can correspond to several template reactions, the task here is to enumerate the different possible transformations according to these templates. Because the RetroRules reaction rules consider only one substrate at a time, some compounds are by purpose omitted: the task here is to complete the predicted reactions by putting back these omitted compounds (mostly cofactors). The node converts each predicted pathway to distinct SBML files.
Question
- How many solutions are found?
- In which file format are the pathways?
- Do these pathways are a good solution?
- We have 9 candidates.
- The pathways are represented in a SBML format.
- It’s a trap! We don’t know if they represent a good solution. We need to evaluate them.
Run the RetroSynthesis Workflow
In this section, you can run the RetroSynthesis Workflow more easily and fastly following these instructions:
Hands-on: Execute the entire workflow in one go.
Import your RetroSynthesis workflow by uploading the workflow file.
- Click on Workflow on the top menu bar of Galaxy. You will see a list of all your workflows.
- Click on the upload icon galaxy-upload at the top-right of the screen
- Provide your workflow
- Option 1: Paste the URL of the workflow into the box labelled “Archived Workflow URL”
- Option 2: Upload the workflow file in the box labelled “Archived Workflow File”
- Click the Import workflow button
- Click on Workflow on the top menu bar of Galaxy. You will see RetroSynthesis workflow.
- Click on the workflow-run (Run workflow) button next to your workflow
- Provide the workflow with the following parameters:
- “Target to produce”: Provide the following Inchi source:
InChI=1S/C40H56/c1-33(2)19-13-23-37(7)27-17-31-39(9)29-15-25-35(5)21-11-12-22-36(6)26-16-30-40(10)32-18-28-38(8)24-14-20-34(3)4/h11-12,15-22,25-32H,13-14,23-24H2,1-10H3/b12-11+,25-15+,26-16+,31-17+,32-18+,35-21+,36-22+,37-27+,38-28+,39-29+,40-30+- “Strain”: Select
Escherichia coli str. K-12 substr. MG1655 (iML1515)SBML model.CommentAll the outputs will be automatically generated and identical to the previous ones.
Conclusion
In this tutorial we produced candidates pathways to produce Lycopene in an Esherichia Coli strain. Three main steps were involved:
- Preprocessing data
- Run Retrosynthesis algorithm with RetroPath2.0
- Enumerate all solutions found by RetroPath2.0
Key points
Get heterologous pathways which produce a compound in a chassis
Frequently Asked Questions
Have questions about this tutorial? Check out the FAQ page for the Synthetic Biology topic to see if your question is listed there. If not, please ask your question on the GTN Gitter Channel or the Galaxy Help ForumReferences
- Delépine, B., T. Duigou, P. Carbonell, and J.-L. Faulon, 2018 RetroPath2.0: A retrosynthesis workflow for metabolic engineers. Metabolic Engineering 45: 158–170. 10.1016/j.ymben.2017.12.002
- Duigou, T., M. du Lac, P. Carbonell, and J.-L. Faulon, 2018 RetroRules: a database of reaction rules for engineering biology. Nucleic Acids Research 47: D1229–D1235. 10.1093/nar/gky940
- Hérisson, J., T. Duigou, M. du Lac, K. Bazi-Kabbaj, M. S. Azad et al., 2022 Galaxy-SynBioCAD: Automated Pipeline for Synthetic Biology Design and Engineering. 10.1101/2022.02.23.481618
Feedback
Did you use this material as an instructor? Feel free to give us feedback on how it went.
Did you use this material as a learner or student? Click the form below to leave feedback.
Citing this Tutorial
- Guillaume Gricourt, Thomas Duigou, Kenza Bazi-Kabbaj, Joan Hérisson, Ioana Popescu, Jean-Loup Faulon, 2022 Generating theoretical possible pathways for the production of Lycopene in E.Coli using Retrosynthesis tools (Galaxy Training Materials). https://training.galaxyproject.org/training-material/topics/synthetic-biology/tutorials/retrosynthesis_analysis/tutorial.html Online; accessed TODAY
- Batut et al., 2018 Community-Driven Data Analysis Training for Biology Cell Systems 10.1016/j.cels.2018.05.012
Congratulations on successfully completing this tutorial!@misc{synthetic-biology-retrosynthesis_analysis, author = "Guillaume Gricourt and Thomas Duigou and Kenza Bazi-Kabbaj and Joan Hérisson and Ioana Popescu and Jean-Loup Faulon", title = "Generating theoretical possible pathways for the production of Lycopene in E.Coli using Retrosynthesis tools (Galaxy Training Materials)", year = "2022", month = "10", day = "18" url = "\url{https://training.galaxyproject.org/training-material/topics/synthetic-biology/tutorials/retrosynthesis_analysis/tutorial.html}", note = "[Online; accessed TODAY]" } @article{Batut_2018, doi = {10.1016/j.cels.2018.05.012}, url = {https://doi.org/10.1016%2Fj.cels.2018.05.012}, year = 2018, month = {jun}, publisher = {Elsevier {BV}}, volume = {6}, number = {6}, pages = {752--758.e1}, author = {B{\'{e}}r{\'{e}}nice Batut and Saskia Hiltemann and Andrea Bagnacani and Dannon Baker and Vivek Bhardwaj and Clemens Blank and Anthony Bretaudeau and Loraine Brillet-Gu{\'{e}}guen and Martin {\v{C}}ech and John Chilton and Dave Clements and Olivia Doppelt-Azeroual and Anika Erxleben and Mallory Ann Freeberg and Simon Gladman and Youri Hoogstrate and Hans-Rudolf Hotz and Torsten Houwaart and Pratik Jagtap and Delphine Larivi{\`{e}}re and Gildas Le Corguill{\'{e}} and Thomas Manke and Fabien Mareuil and Fidel Ram{\'{\i}}rez and Devon Ryan and Florian Christoph Sigloch and Nicola Soranzo and Joachim Wolff and Pavankumar Videm and Markus Wolfien and Aisanjiang Wubuli and Dilmurat Yusuf and James Taylor and Rolf Backofen and Anton Nekrutenko and Björn Grüning}, title = {Community-Driven Data Analysis Training for Biology}, journal = {Cell Systems} }

 Questions:
Questions:
