Chloroplast genome assembly
OverviewQuestions:Objectives:
How can we assemble a chloroplast genome?
Requirements:
Assemble a chloroplast genome from long reads
Polish the assembly with short reads
Annotate the assembly and view
Map reads to the assembly and view
- Introduction to Galaxy Analyses
- Sequence analysis
- Quality Control: slides slides - tutorial hands-on
Time estimation: 2 hoursSupporting Materials:Last modification: Oct 18, 2022
Introduction
What is genome assembly?
Genome assembly is the process of joining together DNA sequencing fragments into longer pieces, ideally up to chromosome lengths.The DNA fragments are produced by DNA sequencing machines, and are called “reads”. These are in lengths of about 150 nucleotides (base pairs), to up to a million+ nucleotides, depending on the sequencing technology used. Currently, most reads are from Illumina (short), PacBio (long) or Oxford Nanopore (long and extra-long).
It is difficult to assemble plant genomes as they are often large (for example, 3,000,000,000 base pairs), have many repeat regions (such as transposons), and may be polyploid. This tutorial shows genome assembly for a smaller data set - the plant chloroplast genome - a single circular chromosome which is typically about 160,000 base pairs. It is thought that the the chloroplast evolved from a cyanobacteria that was living in plant cells.
In this tutorial, we will use a subset of a real data set from sweet potato, from the paper Zhou et al. 2018. To find out more about each of the tools used here, see the tool panel page for a summary and links to more information.
AgendaIn this tutorial we will deal with:
Upload data
Let’s start with uploading the data.
Hands-on: Import the data
Create a new history for this tutorial and give it a proper name
Click the new-history icon at the top of the history panel.
If the new-history is missing:
- Click on the galaxy-gear icon (History options) on the top of the history panel
- Select the option Create New from the menu
- Click on Unnamed history (or the current name of the history) (Click to rename history) at the top of your history panel
- Type the new name
- Press Enter
Import from Zenodo or a data library (ask your instructor):
- FASTQ file with illumina reads:
sweet-potato-chloroplast-illumina-reduced.fastq- FASTQ file with nanopore reads:
sweet-potato-chloroplast-nanopore-reduced.fastq- Note: make sure to import the files with “reduced” in the names, not the ones with “tiny” in the names.
https://zenodo.org/record/3567224/files/sweet-potato-chloroplast-illumina-reduced.fastq https://zenodo.org/record/3567224/files/sweet-potato-chloroplast-nanopore-reduced.fastq
- Copy the link location
Open the Galaxy Upload Manager (galaxy-upload on the top-right of the tool panel)
- Select Paste/Fetch Data
Paste the link into the text field
Press Start
- Close the window
As an alternative to uploading the data from a URL or your computer, the files may also have been made available from a shared data library:
- Go into Shared data (top panel) then Data libraries
- Navigate to the correct folder as indicated by your instructor
- Select the desired files
- Click on the To History button near the top and select as Datasets from the dropdown menu
- In the pop-up window, select the history you want to import the files to (or create a new one)
- Click on Import
Check read quality
We will look at the quality of the nanopore reads.
Hands-on: Check read quality
- Nanoplot Tool: toolshed.g2.bx.psu.edu/repos/iuc/nanoplot/nanoplot/1.28.2+galaxy1 :
- “Select multifile mode”:
batch- “Type of file to work on”:
fastq- “files”: select the
nanopore FASTQ file- View output:
- There are five output files.
- Look at the
HTML reportto learn about the read quality.
QuestionWhat summary statistics would be useful to look at?
This will depend on the aim of your analysis, but usually:
- Sequencing depth (the number of reads covering each base position; also called “coverage”). Higher depth is usually better, but at very high depths it may be better to subsample the reads, as errors can swamp the assembly graph.
- Sequencing quality (the quality score indicates probability of base call being correct). You may trim or filter reads on quality. Phred quality scores are logarithmic: phred quality 10 = 90% chance of base call being correct; phred quality 20 = 99% chance of base call being correct. More detail here.
- Read lengths (read lengths histogram, and reads lengths vs. quality plots). Your analysis or assembly may need reads of a certain length.
Optional further steps:
- Find out the quality of your reads using other tools such as fastp or FastQC.
- To visualize base quality using emoji you can also use FASTQE.
- Run FASTQE for the illumina reads. In the output, look at the mean values (the middle row)
- Repeat FASTQE for the nanopore reads. In the tool settings, increase the maximum read length to 30000.
- To learn more, see the Quality Control tutorial
Assemble reads
We will assemble the long nanopore reads.
Hands-on: Assemble reads
- Flye Tool: toolshed.g2.bx.psu.edu/repos/bgruening/flye/flye/2.6+galaxy0 :
- “Input reads”:
sweet-potato-chloroplast-nanopore-reduced.fastq- “Estimated genome size”:
160000- Leave other settings as default
Re-name the
consensusoutput file toflye-assembly.fasta
- Click on the galaxy-pencil pencil icon for the dataset to edit its attributes
- In the central panel, change the Name field to
flye-assembly.fasta- Click the Save button
- View output:
- There are five output files.
- Note: this tool is heuristic; your results may differ slightly from the results here, and if repeated.
- View the
logfile and scroll to the end to see how many contigs (fragments) were assembled and the length of the assembly.- View the
assembly_infofile to see contig names and lengths.
Hands-on: View the assembly
- Bandage Info Tool: toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_info/0.8.1+galaxy1
- “Graphical Fragment Assembly”: the Flye output file
Graphical Fragment Assembly(not the “assembly_graph” file)- Leave other settings as default
- Bandage Image Tool: toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_image/0.8.1+galaxy2
- “Graphical Fragment Assembly”: the Flye output file
Graphical Fragment Assembly(not the “assembly_graph” file)- “Node length labels”:
Yes- Leave other settings as default
Your assembly graph may look like this:
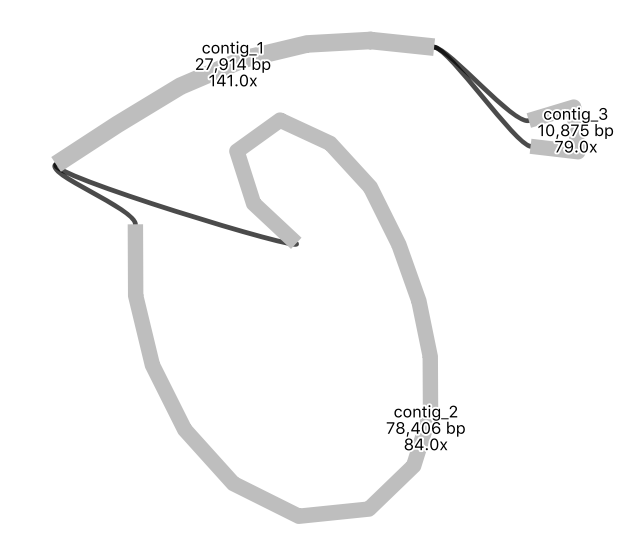
Note: a newer version of the Flye assembly tool now resolves this assembly into a single circle.
QuestionWhat is your interpretation of this assembly graph?
One interpretation is that this represents the typical circular chloroplast structure: There is a long single-copy region (the node of around 78,000 bp), connected to the inverted repeat (a node of around 28,000 bp), connected to the short single-copy region (of around 11,000 bp). In the graph, each end loop is a single-copy region (either long or short) and the centre bar is the collapsed inverted repeat which should have about twice the sequencing depth.
Comment: Further Learning
- Repeat the Flye assembly with different parameters, and/or a filtered read set.
- You can also try repeating the Flye assembly with an earlier version of the tool, to see the difference it makes. In the tool panel for Flye, click on the ‘Versions’ button at the top to change.
- Try an alternative assembly tool, such as Canu or Unicycler.
Polish assembly
Short illumina reads are more accurate the nanopore reads. We will use them to correct errors in the nanopore assembly.
First, we will map the short reads to the assembly and create an alignment file.
Hands-on: Map reads
- Map with BWA-MEM Tool: toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.17.1 :
- “Will you select a reference genome from your history”:
Use a genome from history- “Use the following dataset as the reference sequence”:
flye-assembly.fasta- “Algorithm for constructing the BWT index”:
Auto. Let BWA decide- “Single or Paired-end reads”:
Single- “Select fastq dataset”:
sweet-potato-illumina-reduced.fastq- “Set read groups information?”:
Do not set- “Select analysis mode”:
Simple Illumina mode- Re-name output file:
- Re-name this file
illumina.bam
Next, we will compare the short reads to the assembly, and create a polished (corrected) assembly file.
Hands-on: Polish
- pilon Tool: toolshed.g2.bx.psu.edu/repos/iuc/pilon/pilon/1.20.1 :
- “Source for reference genome used for BAM alignments”:
Use a genome from history- “Select a reference genome”:
flye-assembly.fasta- “Type automatically determined by pilon”:
Yes- “Input BAM file”:
illumina.bam- “Variant calling mode”:
No- “Create changes file”:
Yes- View output:
- What is in the
changesfile?- Rename the fasta output to
polished-assembly.fasta
- Click on the galaxy-pencil pencil icon for the dataset to edit its attributes
- In the central panel, change the Name field to
polished-assembly.fasta- Click the Save button
- Fasta Statistics Tool: toolshed.g2.bx.psu.edu/repos/iuc/fasta_stats/fasta-stats/1.0.1
- Find and run the tool called “Fasta statistics” on both the original flye assembly and the polished version.
QuestionHow does the polished assembly compare to the unpolished assembly?
This will depend on the settings, but as an example: your polished assembly might be about 10-15 Kbp longer. Nanopore reads can have homopolymer deletions - a run of AAAA may be interpreted as AAA - so the more accurate illumina reads may correct these parts of the long-read assembly. In the changes file, there may be a lot of cases showing a supposed deletion (represented by a dot) being corrected to a base.
Optional further steps:
- Run a second round (or more) of Pilon polishing. Keep track of file naming; you will need to generate a new bam file first before each round of Pilon.
- Run an alternative polishing tool, such as Racon. This uses the long reads themselves to correct the long-read (Flye) assembly. It would be better to run this tool on the Flye assembly before running Pilon, rather than after Pilon.
Annotate the assembly
We can now annotate our assembled genome with information about genomic features.
- A chloroplast genome annotation tool is not yet available in Galaxy; for an approximation, here we can use the tool for bacterial genome annotation, Prokka.
Hands-on: Annotate with Prokka
- Prokka Tool: toolshed.g2.bx.psu.edu/repos/crs4/prokka/prokka/1.14.5+galaxy0 with the following parameters (leave everything else unchanged)
- param-file “contigs to annotate”:
polished-assembly.fasta- View output:
- The GFF and GBK files contain all of the information about the features annotated (in different formats.)
- The .txt file contains a summary of the number of features annotated.
- The .faa file contains the protein sequences of the genes annotated.
- The .ffn file contains the nucleotide sequences of the genes annotated.
Alternatively, you might want to use a web-based tool designed for chloroplast genomes.
- One option is the GeSeq tool, described here. Skip this step if you have already used Prokka above.
Hands-on: Annotate with GeSeq
- Download
polished-assembly.fastato your computer (click on the file in your history; then click on the disk icon).- In a new browser tab, go to Chlorobox where we will use the GeSeq tool (Tillich et al. 2017) to annotate our sequence.
- Upload the
fastafile there. Information about how to use the tool is available on the page.- Once the annotation is completed, download the required files.
- In Galaxy, import the annotation
GFF3file.
Now make a JBrowse file to view the annotations (the GFF3 file - produced from either Prokka or GeSeq) under the assembly (the polished-assembly.fasta file).
Hands-on: View annotations
- JBrowse genome browser Tool: toolshed.g2.bx.psu.edu/repos/iuc/jbrowse/jbrowse/1.16.4+galaxy3 :
- “Reference genome to display”:
Use a genome from history
- “Select a reference genome”:
polished-assembly.fasta- “Produce Standalone Instance”:
Yes- “Genetic Code”:
11. The Bacterial, Archaeal and Plant Plastid Code- “JBrowse-in-Galaxy Action”:
New JBrowse instance- “Insert Track Group”
- “Insert Annotation Track”
- “Track Type”:
GFF/GFF3/BED Features- “GFF/GFF3/BED Track Data”: the
GFF3file- Leave the other track features as default
- Re-name output file:
- JBrowse may take a few minutes to run. There is one output file: re-name it
view-annotations- View output:
- Click on the eye icon to view the annotations file.
- Select the right contig to view, in the drop down box.
- Zoom out (with the minus button) until annotations are visible.
Here is an embedded snippet showing JBrowse and the annotations:
View reads
We will look at the original sequencing reads mapped to the genome assembly. In this tutorial, we will import very cut-down read sets so that they are easier to view.
Hands-on: Import cut-down read sets
- Import Tool: upload1 from Zenodo or a data library (ask your instructor):
- FASTQ file with illumina reads:
sweet-potato-chloroplast-illumina-tiny.fastq- FASTQ file with nanopore reads:
sweet-potato-chloroplast-nanopore-tiny.fastq- Note: these are the “tiny” files, not the “reduced” files we imported earlier.
https://zenodo.org/record/3567224/files/sweet-potato-chloroplast-illumina-tiny.fastq https://zenodo.org/record/3567224/files/sweet-potato-chloroplast-nanopore-tiny.fastq
Hands-on: Map the reads to the assembly
- Map the Illumina reads (the new “tiny” dataset) to the
polished-assembly.fasta, the same way we did before, using bwa mem.- This creates one output file: re-name it
illumina-tiny.bam- Map the Nanopore reads (the new “tiny” dataset) to the
polished-assembly.fasta. The settings will be the same, exceptSelect analysis modeshould beNanopore- This creates one output file: re-name it
nanopore-tiny.bam
Hands-on: Visualise mapped reads
- JBrowse genome browser Tool: toolshed.g2.bx.psu.edu/repos/iuc/jbrowse/jbrowse/1.16.4+galaxy3 :
- “Reference genome to display”:
Use a genome from history
- “Select a reference genome”:
polished-assembly.fasta- “Produce Standalone Instance”:
Yes- “Genetic Code”:
11. The Bacterial, Archaeal and Plant Plastid Code- “JBrowse-in-Galaxy Action”:
New JBrowse instance- “Insert Track Group”
- “Insert Annotation Track”
- “Track Type”:
BAM pileups- “BAM track data”:
nanopore-tiny.bam- “Autogenerate SNP track”:
No- Leave the other track features as default
- “Insert Annotation Track”.
- “Track Type”:
BAM pileups- “BAM track data”:
illumina-tiny.bam- “Autogenerate SNP track”:
No- Leave the other track features as default
- Re-name output file:
- JBrowse may take a few minutes to run. There is one output file: re-name it
assembly-and-reads- View output:
- Click on the eye icon to view. (For more room, collapse Galaxy side menus with corner < > signs).
- Make sure the bam files are ticked in the left hand panel.
- Choose a contig in the drop down menu. Zoom in and out with + and - buttons.
Here is an embedded snippet showing JBrowse and the mapped reads:
Question
- What are the differences between the nanopore and the illumina reads?
- What are some reasons that the read coverage may vary across the reference genome?
- Nanopore reads are longer and have a higher error rate.
- There may be lots of reasons for varying read coverage. Some possibilities: In areas of high read coverage: this region may be a collapsed repeat. In areas of low or no coverage: this region may be difficult to sequence; or, this region may be a misassembly.
- To learn more about JBrowse and its features, see the Genomic Data Visualisation with JBrowse tutorial
Repeat with new data
Optional extension exercise
We can assemble another chloroplast genome using sequence data from a different plant species: the snow gum, Eucalyptus pauciflora. This data is from Wang et al. 2018. It is a subset of the original FASTQ read files (Illumina - SRR7153063, Nanopore - SRR7153095).
Hands-on: Assembly and annotation
- Get data: at this Zenodo link, then upload to Galaxy.
- Check reads: Run Nanoplot on the nanopore reads.
- Assemble: Use Flye to assemble the nanopore reads, then get Fasta statistics Note: this may take several hours.
- Polish assembly: Use Pilon to polish the assembly with short Illumina reads. Note: Don’t forget to map these Illumina reads to the assembly first using bwa-mem, then use the resulting
bamfile as input to Pilon.- Annotate: Use the GeSeq tool at Chlorobox or the Prokka tool within Galaxy.
- View annotations:Use JBrowse to view the assembled, annotated genome.
Conclusion
Key points
A chloroplast genome can be assembled with long reads and polished with short reads
The assembly graph is useful to look at and think about genomic structure
We can map raw reads back to the assembly and investigate areas of high or low read coverage
We can view an assembly, its mapped reads, and its annotations in JBrowse
Frequently Asked Questions
Have questions about this tutorial? Check out the tutorial FAQ page or the FAQ page for the Assembly topic to see if your question is listed there. If not, please ask your question on the GTN Gitter Channel or the Galaxy Help ForumReferences
- Tillich, M., P. Lehwark, T. Pellizzer, E. S. Ulbricht-Jones, A. Fischer et al., 2017 GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Research 45: W6–W11. 10.1093/nar/gkx391
- Wang, W., M. Schalamun, A. Morales-Suarez, D. Kainer, B. Schwessinger et al., 2018 Assembly of chloroplast genomes with long- and short-read data: a comparison of approaches using Eucalyptus pauciflora as a test case. BMC Genomics 19: 10.1186/s12864-018-5348-8
- Zhou, C., T. Duarte, R. Silvestre, G. Rossel, R. O. M. Mwanga et al., 2018 Insights into population structure of East African sweetpotato cultivars from hybrid assembly of chloroplast genomes. Gates Open Research 2: 41. 10.12688/gatesopenres.12856.1
Feedback
Did you use this material as an instructor? Feel free to give us feedback on how it went.
Did you use this material as a learner or student? Click the form below to leave feedback.
Citing this Tutorial
- Anna Syme, 2022 Chloroplast genome assembly (Galaxy Training Materials). https://training.galaxyproject.org/training-material/topics/assembly/tutorials/chloroplast-assembly/tutorial.html Online; accessed TODAY
- Batut et al., 2018 Community-Driven Data Analysis Training for Biology Cell Systems 10.1016/j.cels.2018.05.012
Congratulations on successfully completing this tutorial!@misc{assembly-chloroplast-assembly, author = "Anna Syme", title = "Chloroplast genome assembly (Galaxy Training Materials)", year = "2022", month = "10", day = "18" url = "\url{https://training.galaxyproject.org/training-material/topics/assembly/tutorials/chloroplast-assembly/tutorial.html}", note = "[Online; accessed TODAY]" } @article{Batut_2018, doi = {10.1016/j.cels.2018.05.012}, url = {https://doi.org/10.1016%2Fj.cels.2018.05.012}, year = 2018, month = {jun}, publisher = {Elsevier {BV}}, volume = {6}, number = {6}, pages = {752--758.e1}, author = {B{\'{e}}r{\'{e}}nice Batut and Saskia Hiltemann and Andrea Bagnacani and Dannon Baker and Vivek Bhardwaj and Clemens Blank and Anthony Bretaudeau and Loraine Brillet-Gu{\'{e}}guen and Martin {\v{C}}ech and John Chilton and Dave Clements and Olivia Doppelt-Azeroual and Anika Erxleben and Mallory Ann Freeberg and Simon Gladman and Youri Hoogstrate and Hans-Rudolf Hotz and Torsten Houwaart and Pratik Jagtap and Delphine Larivi{\`{e}}re and Gildas Le Corguill{\'{e}} and Thomas Manke and Fabien Mareuil and Fidel Ram{\'{\i}}rez and Devon Ryan and Florian Christoph Sigloch and Nicola Soranzo and Joachim Wolff and Pavankumar Videm and Markus Wolfien and Aisanjiang Wubuli and Dilmurat Yusuf and James Taylor and Rolf Backofen and Anton Nekrutenko and Björn Grüning}, title = {Community-Driven Data Analysis Training for Biology}, journal = {Cell Systems} }
 Questions:
Questions:
