Genome annotation with Maker (short)

OverviewQuestions:Objectives:
How to annotate an eukaryotic genome?
How to evaluate and visualize annotated genomic features?
Requirements:
Load genome into Galaxy
Annotate genome with Maker
Evaluate annotation quality with BUSCO
View annotations in JBrowse
Time estimation: 2 hoursLevel: Intermediate IntermediateSupporting Materials:Last modification: Aug 22, 2022
Introduction
Genome annotation of eukaryotes is a little more complicated than for prokaryotes: eukaryotic genomes are usually larger than prokaryotes, with more genes. The sequences determining the beginning and the end of a gene are generally less conserved than the prokaryotic ones. Many genes also contain introns, and the limits of these introns (acceptor and donor sites) are not highly conserved.
In this tutorial we will use a software tool called Maker Campbell et al. 2014 to annotate the genome sequence of a small eukaryote: Schizosaccharomyces pombe (a yeast).
Maker is able to annotate both prokaryotes and eukaryotes. It works by aligning as many evidences as possible along the genome sequence, and then reconciliating all these signals to determine probable gene structures.
The evidences can be transcript or protein sequences from the same (or closely related) organism. These sequences can come from public databases (like NR or GenBank) or from your own experimental data (transcriptome assembly from an RNASeq experiment for example). Maker is also able to take into account repeated elements.
Maker uses ab-initio predictors (like Augustus or SNAP) to improve its predictions: these software tools are able to make gene structure predictions by analysing only the genome sequence with a statistical model.
In this tutorial you will learn how to perform a genome annotation, and how to evaluate its quality. Finally, you will learn how to use the JBrowse genome browser to visualise the results.
More information about Maker can be found here.
This tutorial was inspired by the MAKER Tutorial for WGS Assembly and Annotation Winter School 2018, don’t hesitate to consult it for more information on Maker, and on how to run it with command line.
Note: Two versions of this tutorial Because this tutorial consists of many steps, we have made two versions of it, one long and one short.
This is the shortened version. We will skip the training of ab-initio predictors and use pre-trained data instead. We will also annotate only the third chromosome of the genome. If you would like to learn how to perform the training steps, please see the longer version of tutorial
In this tutorial, we will cover:
Data upload
To annotate a genome using Maker, you need the following files:
- The genome sequence in fasta format
- A set of transcripts or EST sequences, preferably from the same organism.
- A set of protein sequences, usually from closely related species or from a curated sequence database like UniProt/SwissProt.
Maker will align the transcript and protein sequences on the genome sequence to determine gene positions.
Data upload
Create and name a new history for this tutorial.
Click the new-history icon at the top of the history panel.
If the new-history is missing:
- Click on the galaxy-gear icon (History options) on the top of the history panel
- Select the option Create New from the menu
Import the following files from Zenodo or from the shared data library
https://zenodo.org/api/files/647ad552-19a8-46d9-aad8-f81f56860582/S_pombe_chrIII.fasta https://zenodo.org/api/files/647ad552-19a8-46d9-aad8-f81f56860582/S_pombe_trinity_assembly.fasta https://zenodo.org/api/files/647ad552-19a8-46d9-aad8-f81f56860582/Swissprot_no_S_pombe.fasta https://zenodo.org/api/files/647ad552-19a8-46d9-aad8-f81f56860582/augustus_training_2.tar.gz https://zenodo.org/api/files/647ad552-19a8-46d9-aad8-f81f56860582/snap_training_2.snaphmm
- Copy the link location
Open the Galaxy Upload Manager (galaxy-upload on the top-right of the tool panel)
- Select Paste/Fetch Data
Paste the link into the text field
Press Start
- Close the window
As an alternative to uploading the data from a URL or your computer, the files may also have been made available from a shared data library:
- Go into Shared data (top panel) then Data libraries
- Navigate to the correct folder as indicated by your instructor
- Select the desired files
- Click on the To History button near the top and select as Datasets from the dropdown menu
- In the pop-up window, select the history you want to import the files to (or create a new one)
- Click on Import
- Rename the datasets
Check that the datatype for
augustus_training_2.tar.gzis set toaugustus
- Click on the galaxy-pencil pencil icon for the dataset to edit its attributes
- In the central panel, click on the galaxy-chart-select-data Datatypes tab on the top
- Select
augustus
- tip: you can start typing the datatype into the field to filter the dropdown menu
- Click the Save button
You have the following main datasets:
S_pombe_trinity_assembly.fastacontains EST sequences from S. pombe, assembled from RNASeq data with TrinitySwissprot_no_S_pombe.fastacontains a subset of the SwissProt protein sequence database (public sequences from S. pombe were removed to stay as close as possible to real-life analysis)S_pombe_chrIII.fastacontains only the third chromosome from the full genome of S. pombe
The other datasets will be used later in the tutorial.
Genome quality evaluation
The quality of a genome annotation is highly dependent on the quality of the genome sequences. It is impossible to obtain a good quality annotation with a poorly assembled genome sequence. Annotation tools will have trouble finding genes if the genome sequence is highly fragmented, if it contains chimeric sequences, or if there are a lot of sequencing errors.
Before running the full annotation process, you need first to evaluate the quality of the sequence. It will give you a good idea of what you can expect from it at the end of the annotation.
Get genome sequence statistics
- Fasta Statistics Tool: toolshed.g2.bx.psu.edu/repos/iuc/fasta_stats/fasta-stats/1.0.1 with the following parameters:
- param-file “fasta or multifasta file”: select
S_pombe_chrIII.fastafrom your history
Have a look at the statistics:
num_seq: the number of contigs (or scaffold or chromosomes), compare it to expected chromosome numberslen_min,len_max,len_N50,len_mean,len_median: the distribution of contig sizesnum_bp_not_N: the number of bases that are not N, it should be as close as possible to the total number of bases (num_bp)
These statistics are useful to detect obvious problems in the genome assembly, but it gives no information about the quality of the sequence content. We want to evaluate if the genome sequence contains all the genes we expect to find in the considered species, and if their sequence are correct.
Keep in mind that we are running this tutorial only on the chromosome III instead of the whole genome.
BUSCO (Benchmarking Universal Single-Copy Orthologs) is a tool allowing to answer this question: by comparing genomes from various more or less related species, the authors determined sets of ortholog genes that are present in single copy in (almost) all the species of a clade (Bacteria, Fungi, Plants, Insects, Mammalians, …). Most of these genes are essential for the organism to live, and are expected to be found in any newly sequenced genome from the corresponding clade. Using this data, BUSCO is able to evaluate the proportion of these essential genes (also named BUSCOs) found in a genome sequence or a set of (predicted) transcript or protein sequences. This is a good evaluation of the “completeness” of the genome or annotation.
We will first run this tool on the genome sequence to evaluate its quality.
Run Busco on the genome
- Busco Tool: toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/4.1.4 with the following parameters:
- param-file “Sequences to analyse”: select
S_pombe_chrIII.fastafrom your history- “Mode”:
Genome- “Lineage”:
FungiWe select
Fungias we will annotate the genome of Schizosaccharomyces pombe which belongs to the Fungi kingdom. It is usually better to select the most specific lineage for the species you study. Large lineages (like Metazoa) will consist of fewer genes, but with a strong support. More specific lineages (like Hymenoptera) will have more genes, but with a weaker support (has they are found in fewer genomes).
BUSCO produces three output datasets
- A short summary: summarizes the results of BUSCO (see below)
- A full table: lists all the BUSCOs that were searched for, with the corresponding status (was it found in the genome? how many times? where?)
- A table of missing BUSCOs: this is the list of all genes that were not found in the genome
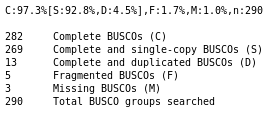
Do you think the genome quality is good enough for performing the annotation?
The genome consists of the expected number of chromosome sequences (1), with very few N, which is the ideal case. As we only analysed chromosome III, many BUSCO genes are missing, but still ~100 are found as complete single copy, and very few are found fragmented, which means that our genome have a good quality, at least on this single chromosome. That’s a very good material to perform an annotation.
Keep in mind that we are running this tutorial only on the chromosome III instead of the whole genome. The BUSCO result will also show a lot of missing genes: it is expected as all the BUSCO genes that are not on the chromosome III cannot be found by the tool.
Maker
Let’s run Maker to predict gene models! Maker will use align ESTs and proteins to the genome, and it will run ab initio predictors (SNAP and Augustus) using pre-trained models for this organism (have a look at the longer version of tutorial to understand how they were trained).
Annotation with Maker
- Maker Tool: toolshed.g2.bx.psu.edu/repos/iuc/maker/maker/2.31.11 with the following parameters:
- param-file “Genome to annotate”: select
S_pombe_chrIII.fastafrom your history- “Organism type”:
Eukaryotic- “Re-annotate using an existing Maker annotation”:
No- In “EST evidences (for best results provide at least one of these)”:
- param-file “ESTs or assembled cDNA”:
S_pombe_trinity_assembly.fasta- In “Protein evidences (for best results provide at least one of these)”:
- param-file “Protein sequences”:
Swissprot_no_S_pombe.fasta- In “Ab-initio gene prediction”:
- “SNAP model”:
snap_training_2.snaphmm- “Prediction with Augustus”:
Run Augustus with a custom prediction model
- param-file “Augustus model”:
augustus_training_2.tar.gz- In “Repeat masking”:
- “Repeat library source”:
Disable repeat masking (not recommended)For this tutorial repeat masking is disabled, which is not the recommended setting. When doing a real-life annotation, you should either use Dfam or provide your own repeats library.
Maker produces three GFF3 datasets:
- The final annotation: the final consensus gene models produced by Maker
- The evidences: the alignments of all the data Maker used to construct the final annotation (ESTs and proteins that we used)
- A GFF3 file containing both the final annotation and the evidences
Annotation statistics
We need now to evaluate this annotation produced by Maker.
First, use the Genome annotation statistics that will compute some general statistics on the annotation.
Get annotation statistics
- Genome annotation statistics Tool: toolshed.g2.bx.psu.edu/repos/iuc/jcvi_gff_stats/jcvi_gff_stats/0.8.4 with the following parameters:
- param-file “Annotation to analyse”:
final annotation(output of Maker Tool: toolshed.g2.bx.psu.edu/repos/iuc/maker/maker/2.31.11 )- “Reference genome”:
Use a genome from history
- param-file “Corresponding genome sequence”: select
S_pombe_chrIII.fastafrom your history
- How many genes where predicted by Maker?
- What is the mean gene locus size of these genes?
- 864 genes
- 1793 bp
Busco
Just as we did for the genome at the beginning, we can use BUSCO to check the quality of this Maker annotation. Instead of looking for known genes in the genome sequence, BUSCO will inspect the transcript sequences of the genes predicted by Maker. This will allow us to see if Maker was able to properly identify the set of genes that Busco found in the genome sequence at the beginning of this tutorial.
First we need to compute all the transcript sequences from the Maker annotation, using GFFread Tool: toolshed.g2.bx.psu.edu/repos/devteam/gffread/gffread/2.2.1.1 . This tool will compute the sequence of each transcript that was predicted by Maker Tool: toolshed.g2.bx.psu.edu/repos/iuc/maker/maker/2.31.11 and write them all in a FASTA file.
Extract transcript sequences
- GFFread Tool: toolshed.g2.bx.psu.edu/repos/devteam/gffread/gffread/2.2.1.1 with the following parameters:
- param-file “Input GFF3 or GTF feature file”:
final annotation(output of Maker Tool: toolshed.g2.bx.psu.edu/repos/iuc/maker/maker/2.31.11 )- “Reference Genome”:
selectS_pombe_chrIII.fastafrom your history
- “Select fasta outputs”:
fasta file with spliced exons for each GFF transcript (-w exons.fa)- “full GFF attribute preservation (all attributes are shown)”:
Yes- “decode url encoded characters within attributes”:
Yes- “warn about duplicate transcript IDs and other potential problems with the given GFF/GTF records”:
Yes
Now run BUSCO with the predicted transcript sequences:
Run BUSCO
- Busco Tool: toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/4.1.4 with the following parameters:
- param-file “Sequences to analyse”:
exons(output of GFFread Tool: toolshed.g2.bx.psu.edu/repos/devteam/gffread/gffread/2.2.1.1 )- “Mode”:
Transcriptome- “Lineage”:
Fungi
How do the BUSCO statistics compare to the ones at the genome level?
128 complete single-copy, 0 duplicated, 10 fragmented, 620 missing. This is in fact better than what BUSCO found in the genome sequence. That means the quality of this annotation is very good (by default BUSCO in genome mode can miss some genes, the advanced options can improve this at the cost of computing time). (Results can be very slightly different in your own history, it’s normal).
Improving gene naming
If you look at the content of the final annotation dataset, you will notice that the gene names are long, complicated, and not very readable. That’s because Maker assign them automatic names based on the way it computed each gene model. We are now going to automatically assign more readable names.
Change gene names
- Map annotation ids Tool: toolshed.g2.bx.psu.edu/repos/iuc/maker_map_ids/maker_map_ids/2.31.11 with the following parameters:
- param-file “Maker annotation where to change ids”:
final annotation(output of Maker Tool: toolshed.g2.bx.psu.edu/repos/iuc/maker/maker/2.31.11 )- “Prefix for ids”:
TEST_- “Justify numeric ids to this length”:
6Genes will be renamed to look like:
TEST_001234. You can replaceTEST_by anything you like, usually an uppercase short prefix.
Look at the generated dataset, it should be much more readable, and ready for an official release.
Visualising the results
With Galaxy, you can visualize the annotation you have generated using JBrowse. This allows you to navigate along the chromosomes of the genome and see the structure of each predicted gene.
Visualize annotations in JBrowse
- JBrowse Tool: toolshed.g2.bx.psu.edu/repos/iuc/jbrowse/jbrowse/1.16.10+galaxy0 with the following parameters:
- “Reference genome to display”:
Use a genome from history
- param-file “Select the reference genome”: select
S_pombe_chrIII.fastafrom your history- “JBrowse-in-Galaxy Action”:
New JBrowse Instance- In “Track Group”:
- Click on “Insert Track Group”:
- In “1: Track Group”:
- “Track Category”:
Maker annotation- In “Annotation Track”:
- Click on “Insert Annotation Track”:
- In “1: Annotation Track”:
“Track Type”:
GFF/GFF3/BED Featuresparam-files “GFF/GFF3/BED Track Data”: select the output of Map annotation ids Tool: toolshed.g2.bx.psu.edu/repos/iuc/maker_map_ids/maker_map_ids/2.31.11
Enable the track on the left side of JBrowse, then navigate along the genome and look at the genes that were predicted by Maker.
Conclusion
Congratulations, you finished this tutorial! You learned how to annotate an eukaryotic genome using Maker, how to evaluate the quality of the annotation, and how to visualize it using the JBrowse genome browser.
What’s next?
After generating your annotation, you will probably want to automatically assign functional annotation to each predicted gene model. You can do it by using Blast, InterProScan, or Blast2GO for example.
An automatic annotation of an eukaryotic genome is rarely perfect. If you inspect some predicted genes, you will probably find some mistakes made by Maker, e.g. wrong exon/intron limits, splitted genes, or merged genes. Setting up a manual curation project using Apollo helps a lot to manually fix these errors. Check out the Apollo tutorial for more details.
Key points
Maker allows to annotate a eukaryotic genome.
BUSCO and JBrowse allow to inspect the quality of an annotation.
Frequently Asked Questions
Have questions about this tutorial? Check out the tutorial FAQ page or the FAQ page for the Genome Annotation topic to see if your question is listed there. If not, please ask your question on the GTN Gitter Channel or the Galaxy Help ForumReferences
- Campbell, M. S., C. Holt, B. Moore, and M. Yandell, 2014 Genome annotation and curation using MAKER and MAKER-P. Current Protocols in Bioinformatics 48: 4–11. 10.1002/0471250953.bi0411s48
Feedback
Did you use this material as an instructor? Feel free to give us feedback on how it went.
Did you use this material as a learner or student? Click the form below to leave feedback.
Citing this Tutorial
- Anthony Bretaudeau, 2022 Genome annotation with Maker (short) (Galaxy Training Materials). https://training.galaxyproject.org/training-material/topics/genome-annotation/tutorials/annotation-with-maker-short/tutorial.html Online; accessed TODAY
- Batut et al., 2018 Community-Driven Data Analysis Training for Biology Cell Systems 10.1016/j.cels.2018.05.012
Congratulations on successfully completing this tutorial!@misc{genome-annotation-annotation-with-maker-short, author = "Anthony Bretaudeau", title = "Genome annotation with Maker (short) (Galaxy Training Materials)", year = "2022", month = "08", day = "22" url = "\url{https://training.galaxyproject.org/training-material/topics/genome-annotation/tutorials/annotation-with-maker-short/tutorial.html}", note = "[Online; accessed TODAY]" } @article{Batut_2018, doi = {10.1016/j.cels.2018.05.012}, url = {https://doi.org/10.1016%2Fj.cels.2018.05.012}, year = 2018, month = {jun}, publisher = {Elsevier {BV}}, volume = {6}, number = {6}, pages = {752--758.e1}, author = {B{\'{e}}r{\'{e}}nice Batut and Saskia Hiltemann and Andrea Bagnacani and Dannon Baker and Vivek Bhardwaj and Clemens Blank and Anthony Bretaudeau and Loraine Brillet-Gu{\'{e}}guen and Martin {\v{C}}ech and John Chilton and Dave Clements and Olivia Doppelt-Azeroual and Anika Erxleben and Mallory Ann Freeberg and Simon Gladman and Youri Hoogstrate and Hans-Rudolf Hotz and Torsten Houwaart and Pratik Jagtap and Delphine Larivi{\`{e}}re and Gildas Le Corguill{\'{e}} and Thomas Manke and Fabien Mareuil and Fidel Ram{\'{\i}}rez and Devon Ryan and Florian Christoph Sigloch and Nicola Soranzo and Joachim Wolff and Pavankumar Videm and Markus Wolfien and Aisanjiang Wubuli and Dilmurat Yusuf and James Taylor and Rolf Backofen and Anton Nekrutenko and Björn Grüning}, title = {Community-Driven Data Analysis Training for Biology}, journal = {Cell Systems} }
 Questions:
Questions:
