name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/genome-annotation" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a class="nav-link" href="https://github.com/galaxyproject/training-material/edit/main/topics/genome-annotation/tutorials/apollo/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/gga.png" alt="page logo" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/gga.png" alt="page logo" class="cover-logo" /> # Refining Genome Annotations with Apollo <div markdown="0"> <div class="contributors-line"> Authors: <a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-abretaud"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/abretaud?s=27" alt="Avatar">Anthony Bretaudeau</a> <a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-hexylena"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/hexylena?s=27" alt="Avatar">Helena Rasche</a> <a href="/training-material/hall-of-fame/nathandunn/" class="contributor-badge contributor-nathandunn"><img src="https://avatars.githubusercontent.com/nathandunn?s=27" alt="Avatar">Nathan Dunn</a> <a href="/training-material/hall-of-fame/mboudet/" class="contributor-badge contributor-mboudet"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/mboudet?s=27" alt="Avatar">Mateo Boudet</a> <a href="/training-material/hall-of-fame/erasmusplus/" class="contributor-badge contributor-erasmusplus"><img src="https://avatars.githubusercontent.com/erasmusplus?s=27" alt="Avatar">Erasmus+ Programme</a> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"><i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: Sep 28, 2022</div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 6em;"> <i class="far fa-play-circle" aria-hidden="true"></i><span class="visually-hidden">video-slides</span> <a href="/training-material/videos/watch.html?v=/genome-annotation/tutorials/apollo/slides">Video slides</a> | <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ## Requirements Before diving into this slide deck, we recommend you to have a look at: - [Introduction to Galaxy Analyses](/training-material/topics/introduction) --- ### <i class="far fa-question-circle" aria-hidden="true"></i><span class="visually-hidden">question</span> Questions - How to visualize your genome after automated annotations have been performed? - How to manually annotate genome after automated annotations have been performed? - How to evaluate and visualize annotated genomic features? - How do I collaborate when doing genome annotation? --- ### <i class="fas fa-bullseye" aria-hidden="true"></i><span class="visually-hidden">objectives</span> Objectives - Load a genome into Galaxy - View annotations in JBrowse - Learn how to load JBrowse data into Apollo - Learn how to manually refine genome annotations within Apollo - Export refined genome annotations --- ### Genome annotation - Classifies and describes parts of the genome sequence - Focusing on genes: their positions, names and functions - Automatic methods to annotate a newly sequenced and assembled genome - Prokaryotes: [Prokka](/training-material/topics/genome-annotation/tutorials/annotation-with-prokka/tutorial.html), Augustus - Eukaryotes: [Maker](/training-material/topics/genome-annotation/tutorials/annotation-with-maker/tutorial.html), Braker2, Augustus .image-50[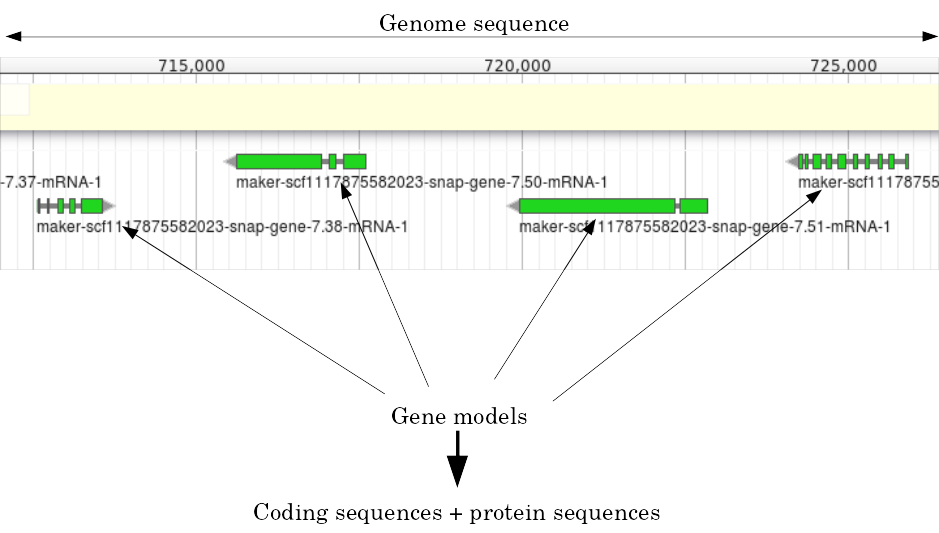] ??? - Annotating a genome means positioning features along the sequence of a genome. - Here we focus on genes. - We are interested in their positions, and functional annotation like their name of their function. - Many software tools and pipelines exist to automatically perform the annotation of a new genome sequence. - For example Prokka or Maker are widely used, and tutorials are available on the GTN to learn how to use them. --- ### Automatic annotations are not perfect .pull-left[ - Assembly errors can cause fragmented annotations - Limited and/or noisy evidence coverage makes precise identification difficult ] .pull-right[.image-40[]] ??? - Unfortunately, automatic annotations are rarely perfect. - Assembly errors can cause fragmented annotations. - Annotation tools have difficulties annotating genes when evidences like RNASeq are lacking or too noisy. --- ### Manual curation refines genome annotations - Make use of the researcher's expertise - Include additional analysis results - Integrate all underlying evidence - A Human finds problems algorithms can't .pull-left[.image-90[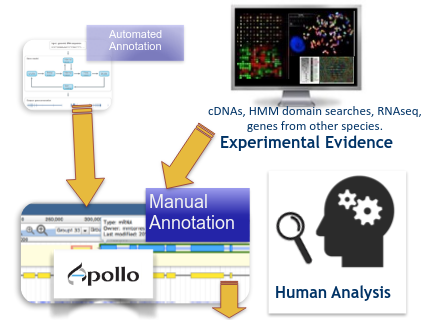]] .pull-right[.image-40[]] ??? - Performing a manual curation step after automatic annotation allows to improve annotation quality. - It is a way to make use of a researcher's expertise, and to integrate more knowledge to improve gene structures and functional annotation. --- ### Apollo "Google Docs for Genome Annotation" .image-50[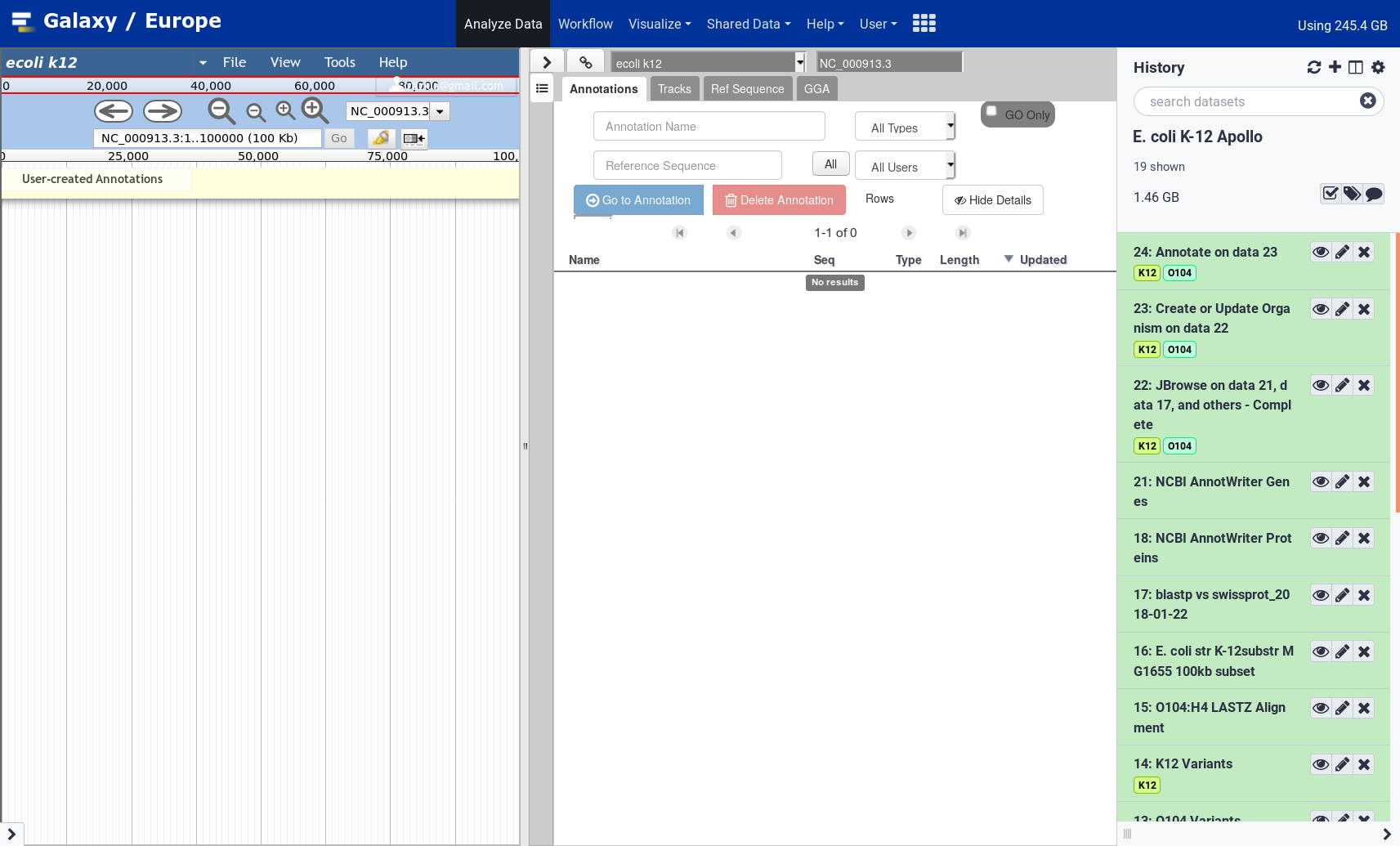] Collaborative web application to edit annotation Both structural *and* functional annotation Based on JBrowse: JBrowse is for visualising only, Apollo allows to edit ??? - Apollo is a web application specifically designed for the manual curation of annotations. - You can see Apollo as the "Google Docs" of annotation. - It is a multi-user, real time, collaborative environment. - Users can easily change the structure of genes, and add functional annotation like gene names of Gene Ontology terms. - Is is based on JBrowse. - JBrowse is for visualising genomes, while Apollo adds the editing capabilities. --- ### Apollo - Modify gene structures (based on evidence tracks) - Add functional annotation .pull-left[.image-90[]] .pull-right[.image-90[]] ??? - Within Apollo, you can easily change the structure of genes, based on evidence tracks. - Apollo offers an interactive interface to help you add functional annotation. --- ### Apollo Multi user environment: users can login, and make/see changes in realtime Full history of changes, reverting is possible 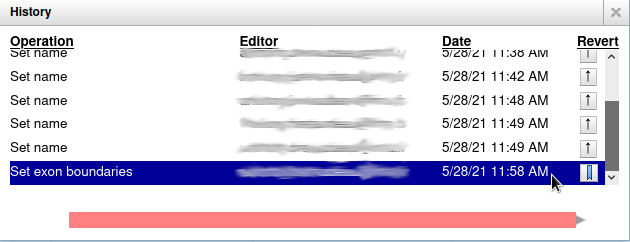 ??? - Apollo is a multi-user environment, which means multiple users can login at the same time, make changes, and see changes of other users in real time. - Apollo keep track of every action performed on each gene in a history. You can revert any change at any time. --- ### Apollo in Galaxy Integrate Apollo in your whole analysis project in Galaxy 1. Run assembly 2. Align evidences along the genome (RNASeq, CDS, proteins) 3. Run automatic annotation pipeline 4. Create a JBrowse instance 5. Load the JBrowse instance into Apollo 6. Use Apollo to manually curate annotation with collaborators 7. Export back the curated annotation into Galaxy 8. Continue analysing in Galaxy Available on [https://usegalaxy.eu](https://usegalaxy.eu) ??? - Apollo is now tightly integrated with Galaxy, which allow to integrate the manual curation process in a complete Galaxy-based analysis workflow. - Within Galaxy, you can start by assembling a new genome, then aligning as many evidence tracks as possible (like RNASeq data). - You can then continue with an automatic annotation step, with Prokka or Maker for example. - Then you can generate a JBrowse instance representing all the data you have generated, and load it into Apollo. - Apollo let's you perform the manual curation, in collaboration with any other colleague you invite to help you. - Finally when you are satisfied with your changes, you can export them to your Galaxy history, and continue other analyses. - At any time later you can come back to Apollo to continue improving the annotation, based on new experiment results. --- ### Apollo in Galaxy: collaborating Invite any other Galaxy user to your curation project 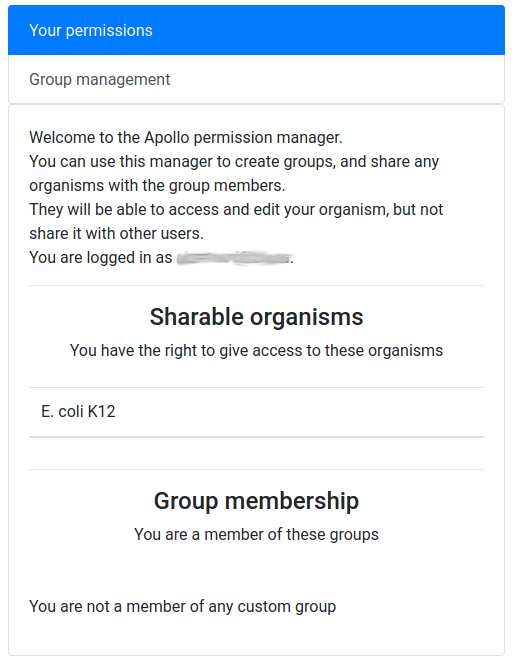 ??? - By default you are the only one having access to genomes you load into Apollo. - A specific module of the Apollo-Galaxy bridge allows you to open the access to a genome to any other Galaxy user. --- <div id="hands-on-hands-on" class="box-title" aria-label="hands-on box: Hands on!"><i class="fas fa-pencil-alt" aria-hidden="true"></i><span class="visually-hidden"></span> Hands-on: Hands on!</div> Time to follow the [Apollo tutorial](/training-material/topics/genome-annotation/tutorials/apollo/tutorial.html)! ??? - You can now follow the Apollo tutorial to learn how to use it in real life. --- ### <i class="fas fa-key" aria-hidden="true"></i><span class="visually-hidden">keypoints</span> Key points - Apollo is the Google Docs of the genome annotation world, real-time collaborative genome annotation. - Apollo allows a group to view and manually refine predicted genome annotations - Use Apollo to edit annotations within your group. - Export manual annotations as GFF3. --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> Authors: <a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-abretaud"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/abretaud?s=27" alt="Avatar">Anthony Bretaudeau</a> <a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-hexylena"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/hexylena?s=27" alt="Avatar">Helena Rasche</a> <a href="/training-material/hall-of-fame/nathandunn/" class="contributor-badge contributor-nathandunn"><img src="https://avatars.githubusercontent.com/nathandunn?s=27" alt="Avatar">Nathan Dunn</a> <a href="/training-material/hall-of-fame/mboudet/" class="contributor-badge contributor-mboudet"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/mboudet?s=27" alt="Avatar">Mateo Boudet</a> <a href="/training-material/hall-of-fame/erasmusplus/" class="contributor-badge contributor-erasmusplus"><img src="https://avatars.githubusercontent.com/erasmusplus?s=27" alt="Avatar">Erasmus+ Programme</a> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/gga.png" alt="page logo" style="height: 100px;"/> </div> <a rel="license" href="https://creativecommons.org/licenses/by/4.0/"> This material is licensed under the Creative Commons Attribution 4.0 International License</a>.