Submitting SARS-CoV-2 sequences to ENA
Contributors
Authors:
 Miguel Roncoroni
Miguel Roncoroni
Objectives
Introduce the European Nucleotide Archive (ENA)
Learn the requirements to submit raw SARS-CoV-2 sequences to ENA in Galaxy
Overview ENA’s metadata model and how metadata objects are linked
last_modification Last modification: Aug 10, 2021
The European Nucleotide Archive
.pull-left[
.left[ ENA is:
- a FAIR and Open repository for sequence data (reads, assemblies, annotations)
- part of the International Nucleotide Sequence Database Collaboration (INSDC) with NCBI and DDJB
- the COVID-19 data portal repository for SARS-CoV-2 sequences
]
]
.pull-right[

The European Nucleotide Archive and INSDC ]
SARS-CoV-2 sequences
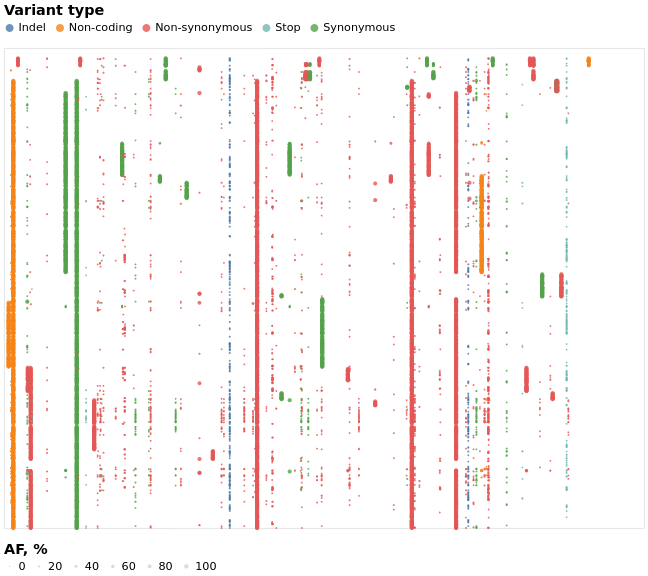
.left[ Why is raw SARS-CoV-2 sequence data important?
- Allows reuse of data and reproducibility of analysis
- Enables discovery of minor allelic variants and intrahost variation
]
.image-40[
 ]
]
.reduce70[Minor allelic-variants can be used to detect intrahost variation. From Maier et al., 2021 doi.org/10.1101/2021.03.25.437046]
Submitting reads with Galaxy
.left[ Why use Galaxy to submit to ENA?
- intuitive graphical user interface (GUI)
- simple metadata input via a template spreadsheet or interactively
- no bioinformatics skills needed
]
.image-75[
 ]
]
Submission overview
.image-100[
 ]
—
]
—
What you need
.left[ Data:
- compressed fastq format (*.fastq.gz, *.fastq.bz2)
- human traces removed (tutorial)
Metadata:
- interactive metadata input (for a few submissions) or;
- metadata template spreadsheet (for bulk submissions)
Credentials:
- ENA Webin credentials in your Galaxy user information
]
.left[
 ]
—
]
—
Metadata
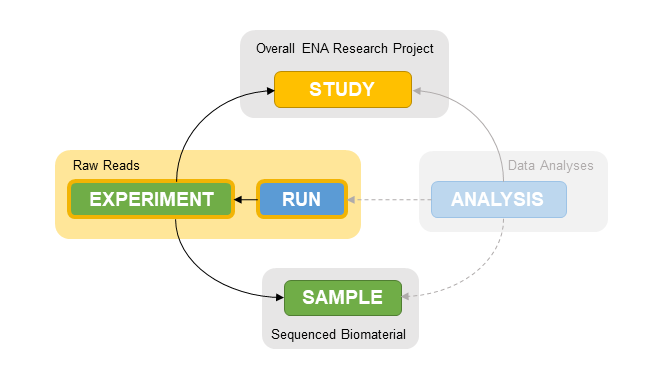
.left[ For the submission of SARS-CoV-2 reads ENA’s metadata model requires:
- study, sample, experiment and run information
- additional information for viral samples (viral checklist)
]
Metadata
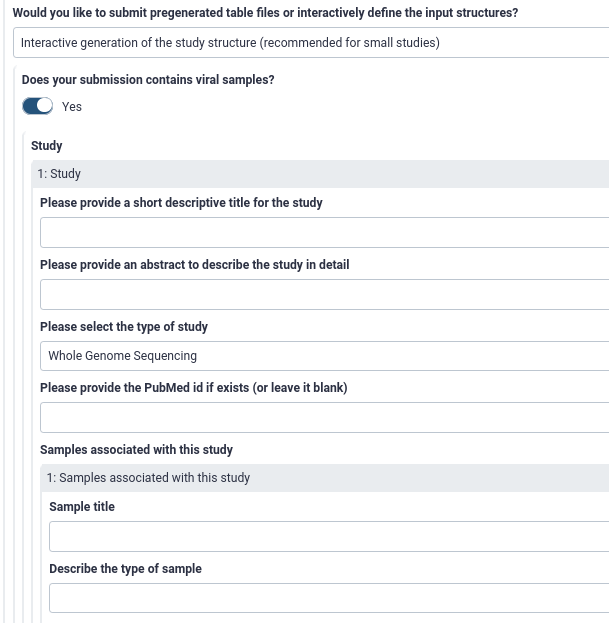
.left[
Interactive metadata input in Galaxy:
]
 —
—
Metadata
.left[ Metadata template spreadsheet:
- one sheet each for study, sample, experiment and run
- built-in controlled vocabulary
]
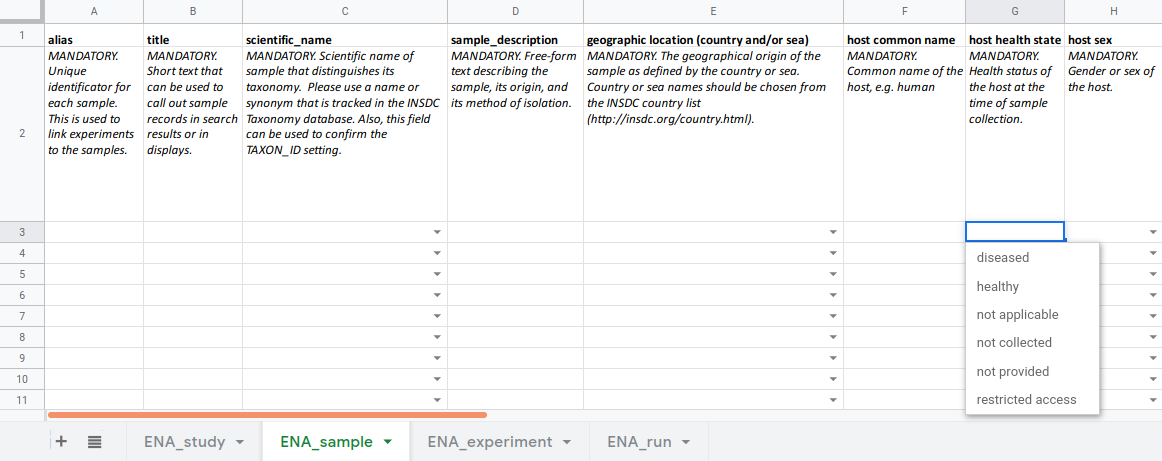 —
—
Metadata
.left[
- Different metadata objects are linked using Aliases
- Aliases must be unique
]
 —
—
Aliases
.left[ Aliases link metadata objects:
- Experiments are linked to Study and Samples
- Runs are linked to Experiments
]
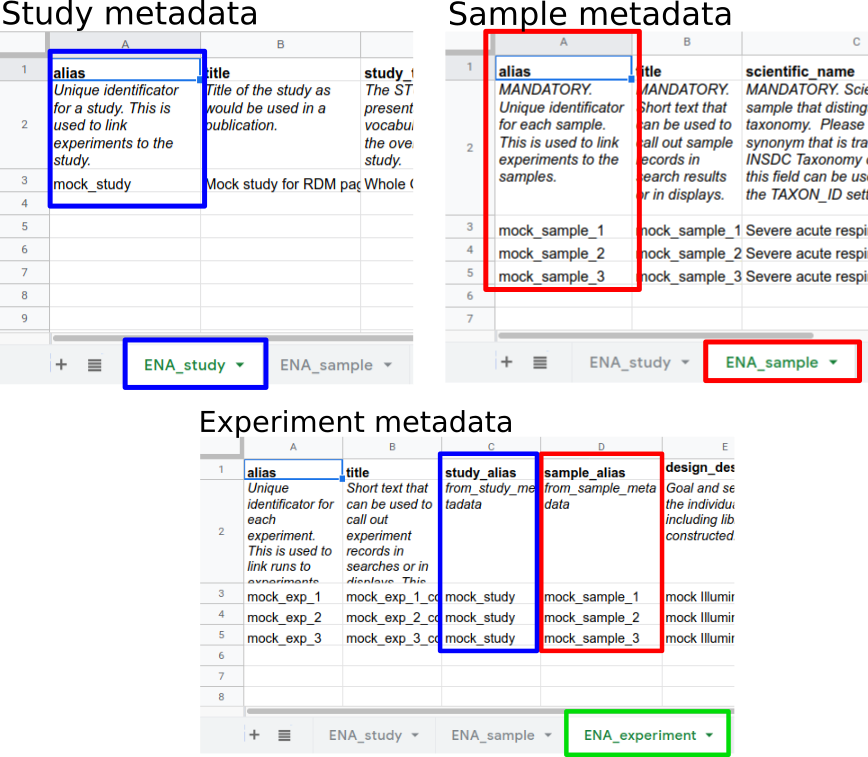 —
—
Aliases
.left[ Aliases link metadata objects:
- Experiments are linked to Study and Samples
- Runs are linked to Experiments
]
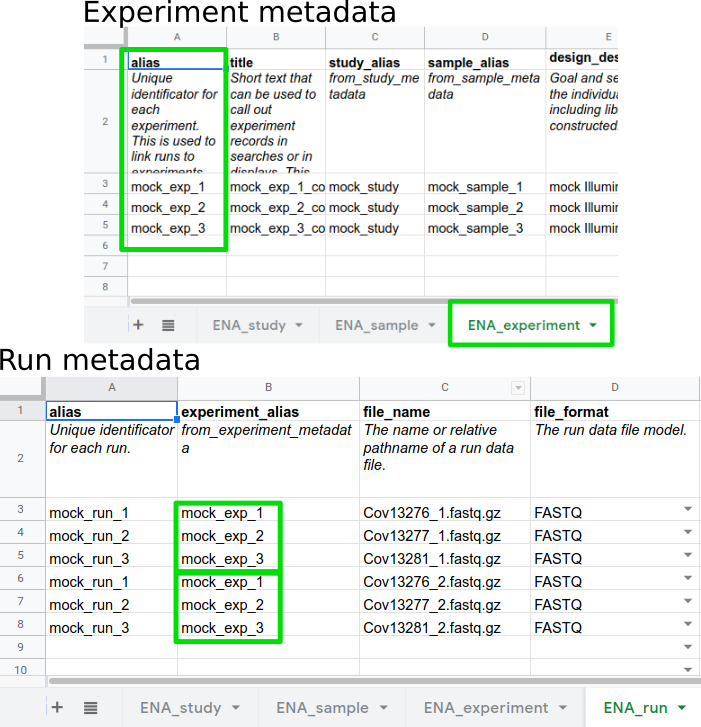 —
—
Aliases
.left[ Aliases link metadata to data:
- Data (filename.fastq.gz) is linked to Run Alias
]
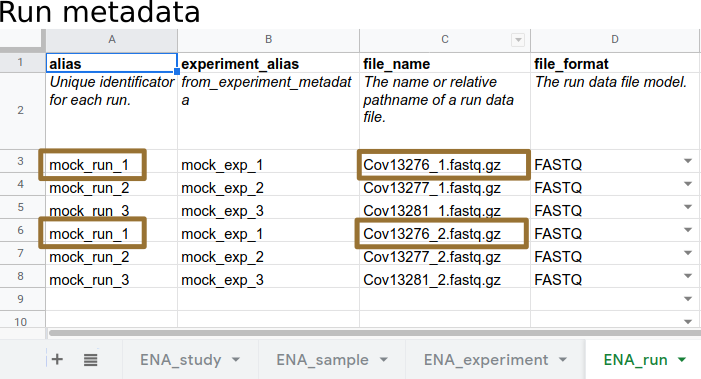 —
—
Key Points
- ENA is a FAIR data repository for SARS-CoV-2 raw and assembled nucleotide data
- You can easily submit reads to ENA using Galaxy's ENA upload tool (GUI, no bioinformatic skills needed)
Thank you!
This material is the result of a collaborative work. Thanks to the Galaxy Training Network and all the contributors! This material is licensed under the Creative Commons Attribution 4.0 International License.
This material is licensed under the Creative Commons Attribution 4.0 International License.