name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/dev" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a class="nav-link" href="https://github.com/galaxyproject/training-material/edit/main/topics/dev/tutorials/tool-integration/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTN.png" alt="Galaxy Training Network" class="cover-logo"/> # Tool development and integration into Galaxy <div markdown="0"> <div class="contributors-line"> Authors: <a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-shiltemann"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/shiltemann?s=27" alt="Avatar">Saskia Hiltemann</a> <a href="/training-material/hall-of-fame/bebatut/" class="contributor-badge contributor-bebatut"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/bebatut?s=27" alt="Avatar">Bérénice Batut</a> <a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-abretaud"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/abretaud?s=27" alt="Avatar">Anthony Bretaudeau</a> <a href="/training-material/hall-of-fame/jmchilton/" class="contributor-badge contributor-jmchilton"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/jmchilton?s=27" alt="Avatar">John Chilton</a> <a href="/training-material/hall-of-fame/nsoranzo/" class="contributor-badge contributor-nsoranzo"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/nsoranzo?s=27" alt="Avatar">Nicola Soranzo</a> <a href="/training-material/hall-of-fame/bgruening/" class="contributor-badge contributor-bgruening"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/bgruening?s=27" alt="Avatar">Björn Grüning</a> <a href="/training-material/hall-of-fame/lecorguille/" class="contributor-badge contributor-lecorguille"><img src="https://avatars.githubusercontent.com/lecorguille?s=27" alt="Avatar">Gildas Le Corguillé</a> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"><i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: Jul 10, 2021</div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 6em;"> <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ### <i class="far fa-question-circle" aria-hidden="true"></i><span class="visually-hidden">question</span> Questions - What is a tool for Galaxy? - How to write a best-practice tool? - How to deal with the tool environment? --- ### <i class="fas fa-bullseye" aria-hidden="true"></i><span class="visually-hidden">objectives</span> Objectives - Learn what a tool is and its structure - Use the Planemo utilities to develop a tool - Deal with the dependencies - Write functional tests - Make a tool ready for publishing in a ToolShed --- # Galaxy tools --- ## Tools in the Galaxy UI 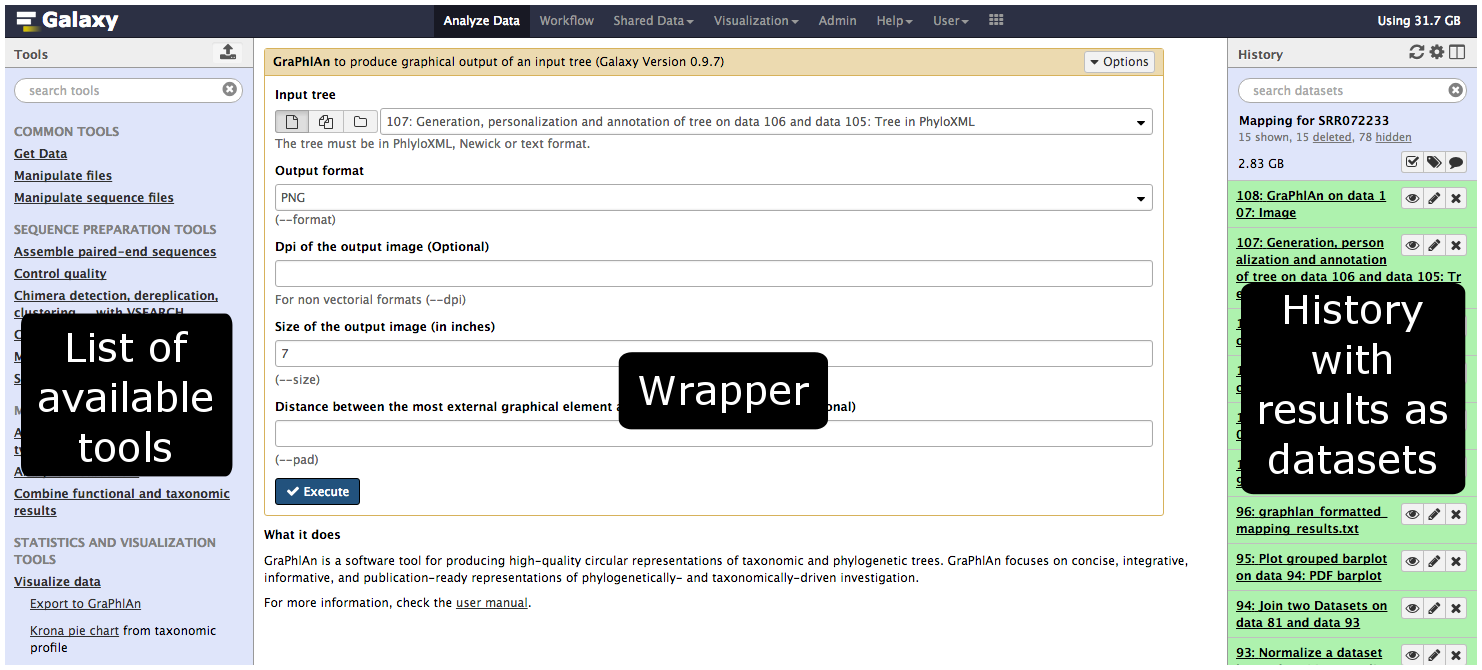 --- ## Galaxy tool / wrapper 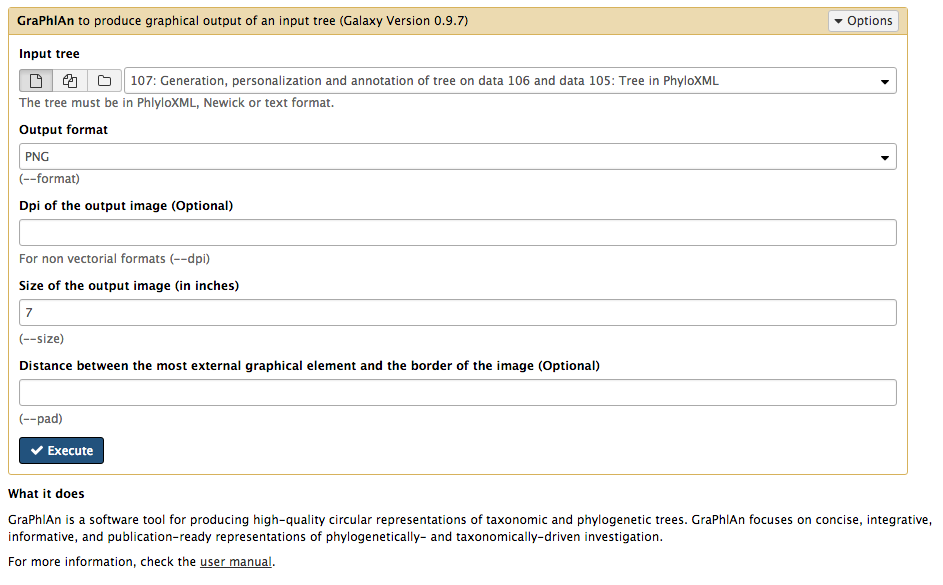 ```bash graphlan.py --format "png" --size 7 input_tree.txt png_image.png ``` --- class: left ## So what is a tool? Link between the Galaxy UI and the underlying tool: - Description of the user interface - How to invoke the tool - Which files and options to pass - Which files the tool will produce as output --- ## Tool execution 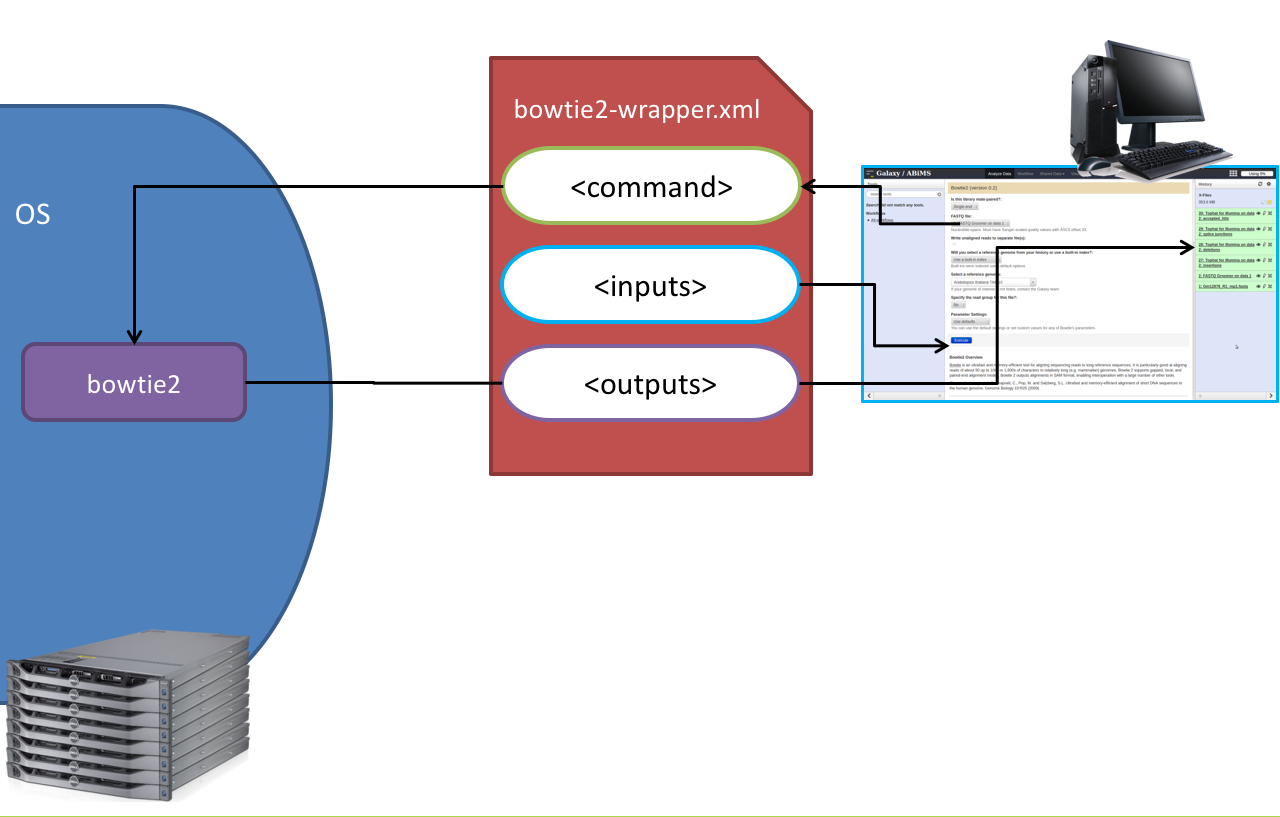 ??? 1. `<inputs>` (datasets and parameters) specified in the tool XML are exposed in the Galaxy tool UI 2. When the user fills the form and click the `Execute` button, Galaxy fills the `<command>` template in the XML with the inputs entered by the user and execute the Cheetah code, producing a script as output 3. Galaxy creates a job for the generated script and executes it somewhere (bowtie2 is run in this case) 4. Some (not necessarily all) output files become new history datasets, as specified in the `<outputs>` XML tag set --- ## Tool execution 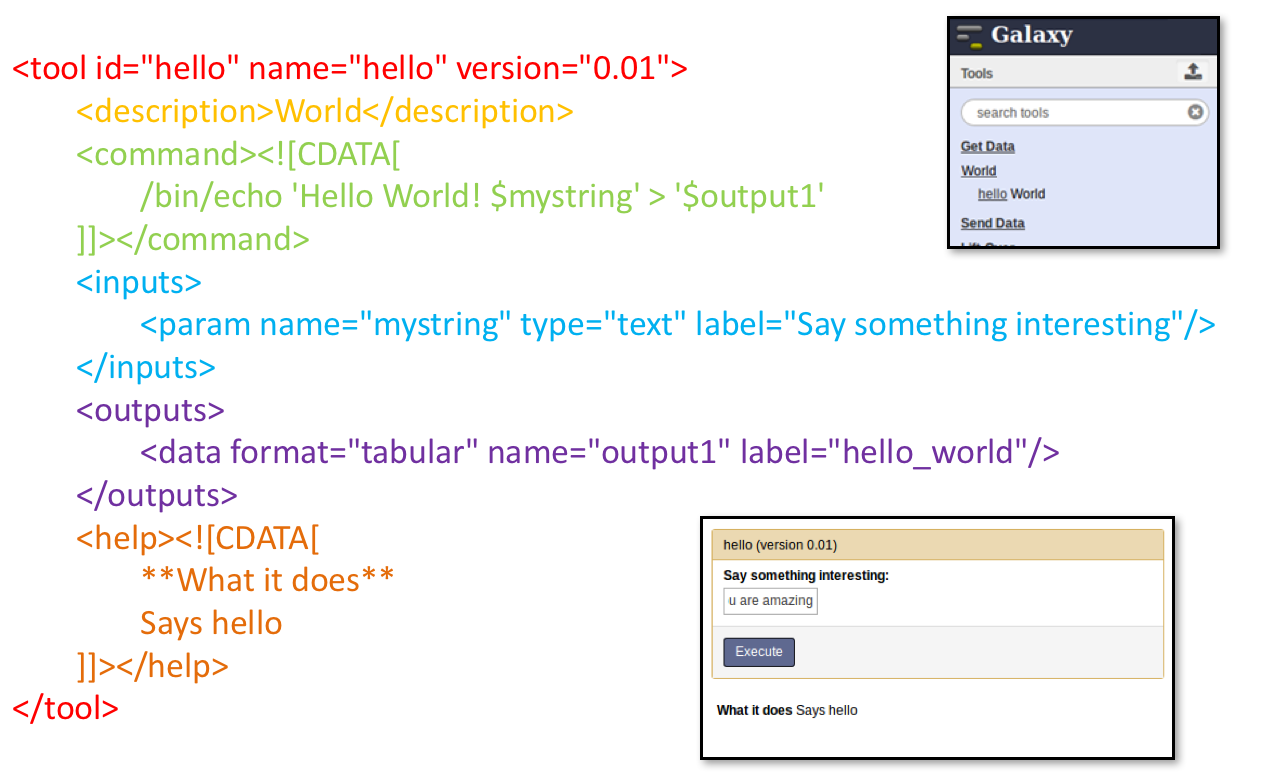 ??? - CDATA tags are used to prevent the interpretation of ampersands and less-than signs as XML special characters - The tool name and description are combined in the left panel (tool menu), keep them short! --- ## Tool execution 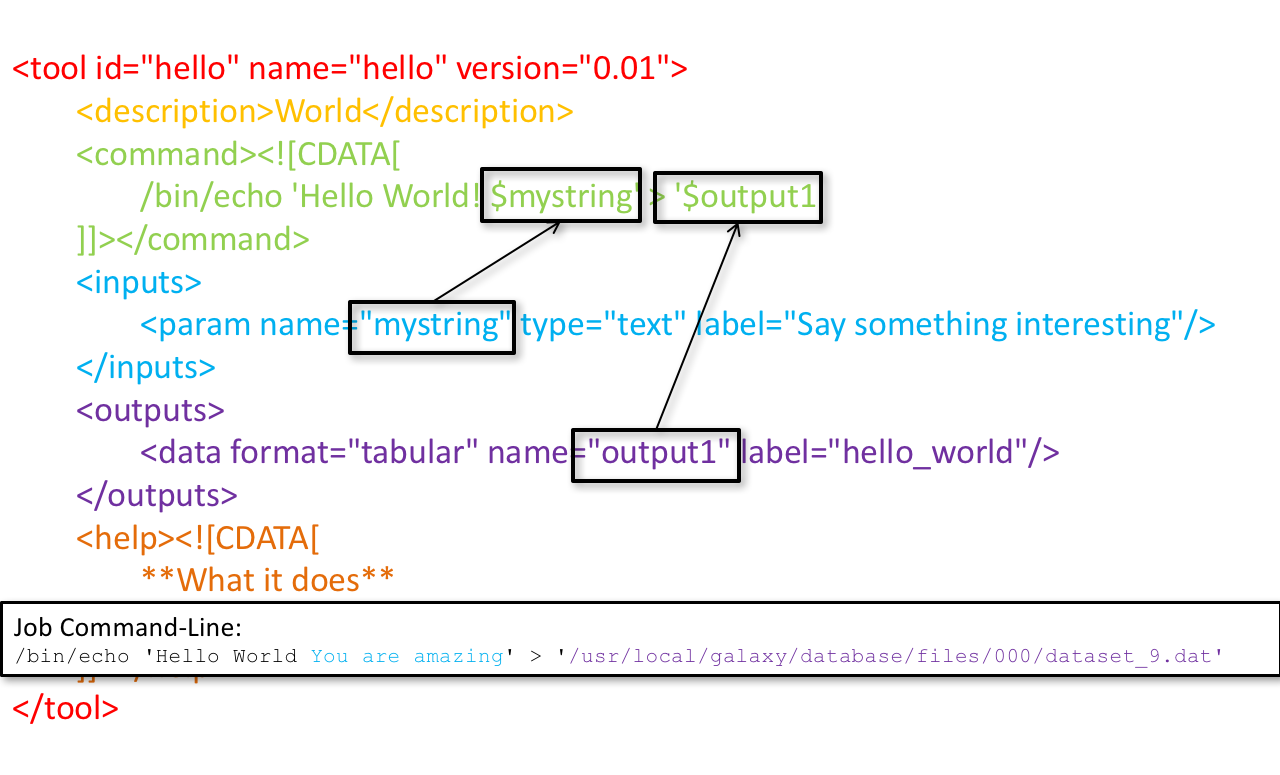 --- ## Tool execution 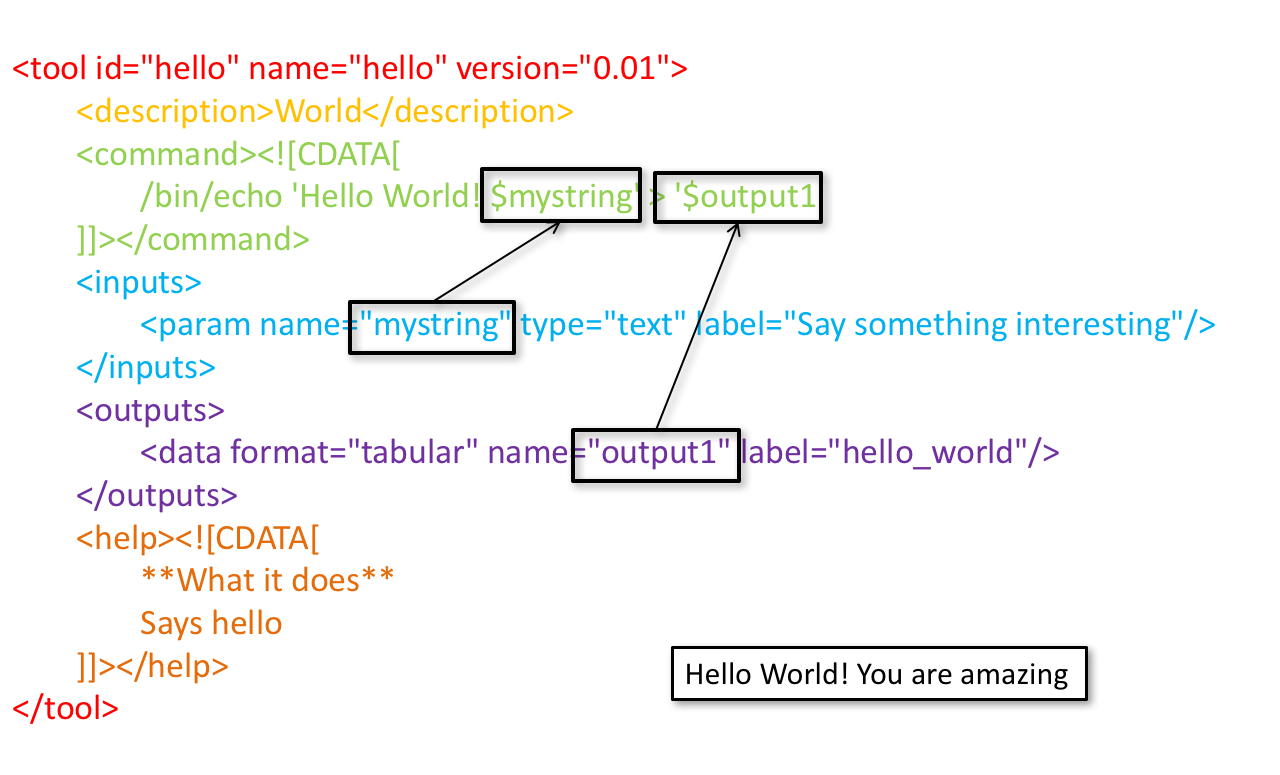 --- ## Tool XML Galaxy tool XML format is formally defined in a XML Schema Definition (XSD), used to generate the corresponding [online documentation](https://docs.galaxyproject.org/en/latest/dev/schema.html) --- ## `command` How to invoke the tool? ```xml <requirements> <requirement type="package" version="1.1.3">graphlan</requirement> </requirements> <command><![CDATA[ graphlan.py --format $format ... ]]></command> ``` If the script is provided with the tool xml: ```xml <requirements> <requirement type="package" version="2.7">python</requirement> </requirements> <command><![CDATA[ python '$__tool_directory__/graphlan.py' --format $format ... ]]></command> ``` ??? - In the first case, `graphlan.py` is expected to be on the PATH and executable when the job executes. This is usually accomplished by specifying some `<requirement/>` tags. - In the second case, `$__tool_directory__` is a special variable which is substituted by Galaxy with the directory where the tool XML is --- ## `inputs` > `param` to `command` Parameters are directly linked to variables in `<command>` ```xml <command><![CDATA[ graphlan.py ... #if str($dpi): --dpi $dpi #end if '$input_tree' ... ]]></command> <inputs> <param name="input_tree" type="data" label="..."/> <param argument="--dpi" type="integer" optional="true" label="..." help="For non vectorial formats" /> </inputs> ``` - The `#if ... #end if` syntax comes from the [Cheetah](https://cheetahtemplate.org/) template language, which has a Python-like syntax ??? - `name` or `argument` identifies a parameter (details of `argument` later) - Parameters can have different types (`data`, `data_collection`, `integer`, `float`, `text`, `select`, `boolean`, `color`, `data_column`,...) --- ## `inputs` > `param` > `data`  ```xml <param name="..." type="data" format="txt" label="..." help="..." /> ``` .footnote[[List of possible formats](https://github.com/galaxyproject/galaxy/blob/dev/config/datatypes_conf.xml.sample)] --- ## `inputs` > `param` > `integer` | `float`  ```xml <param name="..." type="integer" value="7" label="..." help="..."/> ```  ```xml <param name="..." type="float" min="0" max="10" value="1" label="..." help="..."/> ``` ??? - In the first case, Galaxy creates a text box which accepts only integer values - In the second case, since *both* `min` and `max` are specified, a slider is shown in addition --- ## `inputs` > `param` > `text`  ```xml <param name="..." type="text" value="..." label="..." help="..."/> ``` --- ## `inputs` > `param` > `select` 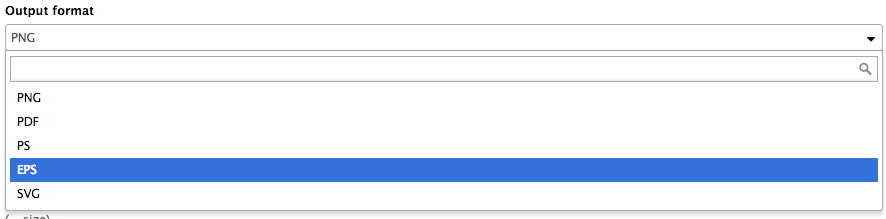 ```xml <param name="..." type="select" label="..." help="..."> <option value="png" selected="true">PNG</option> <option value="pdf">PDF</option> <option value="ps">PS</option> <option value="eps">EPS</option> <option value="svg">SVG</option> </param> ``` If no `option` has `selected="true"`, the first one is selected by default. --- ## `inputs` > `param` > `select`  ```xml <param name="..." type="select" display="radio" label="..." help="..."> <option value="min" selected="true">Minimum</option> <option value="mean">Mean</option> <option value="max">Max</option> <option value="sum">Sum</option> </param> ``` --- ## `inputs` > `param` > `select` 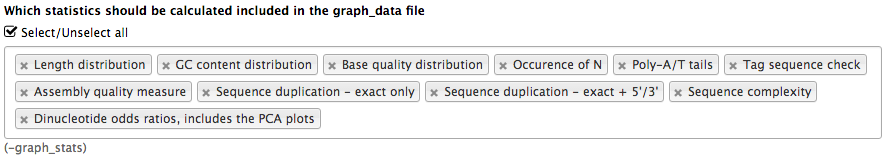 ```xml <param name="..." type="select" multiple="true" label="..." help="..."> <option value="ld" selected="true">Length distribution</option> <option value="gc" selected="true">GC content distribution</option> ... </param> ``` --- ## `inputs` > `param` > `boolean`  ```xml <param name="..." type="boolean" checked="false" truevalue="--log" falsevalue="" label="..." help="..." /> ``` --- class: reduce70 ## `inputs` > `param` > `conditional` .image-75[  ] ```xml <command><![CDATA[ #if $fastq_input.selector == 'paired': '$fastq_input.input1' '$fastq_input.input2' #else: '$fastq_input.input' #end if ]]></command> <inputs> <conditional name="fastq_input"> <param name="selector" type="select" label="Single or paired-end reads?"> <option value="paired">Paired-end</option> <option value="single">Single-end</option> </param> <when value="paired"> <param name="input1" type="data" format="fastq" label="Forward reads" /> <param name="input2" type="data" format="fastq" label="Reverse reads" /> </when> <when value="single"> <param name="input" type="data" format="fastq" label="Single reads" /> </when> </conditional> </inputs> ``` --- class: reduce70 ## `inputs` > `param` > `repeat`  ```xml <command><![CDATA[ #for $i, $s in enumerate($series): rank_of_series=$i input_path=${s.input} x_column=${s.xcol} #end for ]]></command> <inputs> <repeat name="series" title="Series"> <param name="input" type="data" format="tabular" label="Dataset"/> <param name="xcol" type="data_column" data_ref="input" label="Column for x axis"/> </repeat> </inputs> ``` ??? It makes sense to use a `<repeat>` block only if it contains multiple related parameters, otherwise adding `multiple="true"` is preferable. --- ## `outputs` Which files the tool will produce as output?  --- ## `outputs` ```xml <outputs> <data name="tree" format="txt" label="${tool.name} on ${on_string}: Tree" /> <data name="annotation" format="txt" label="${tool.name} on ${on_string}: Annotation" /> </outputs> ``` ??? `${tool.name} on ${on_string}` is the default output label, need to modify this if the tool generates more than 1 output --- ## `outputs` > `filter` Output is collected only if the `filter` evaluates to True ```xml <inputs> <param type="select" name="format" label="Output format"> <option value="png">PNG</option> <option value="pdf">PDF</option> </param> </inputs> <outputs> <data name="png_output" format="png" label="${tool.name} on ${on_string}: PNG"> <filter>format == "png"</filter> </data> <data name="pdf_output" format="pdf" label="${tool.name} on ${on_string}: PDF"> <filter>format == "pdf"</filter> </data> </outputs> ``` ??? N.B. If the filter expression raises an Exception, the dataset will NOT be filtered out --- ## `detect_errors` Legacy tools (i.e. with `profile` unspecified or less than 16.04) by default fail only if the tool writes to stderr Non-legacy tools by default fail if the tool exit code is not 0, which is equivalent to specify: ```xml <command detect_errors="exit_code"> ... </command> ``` To fail if either the tool exit code is not 0 or "Exception:"/"Error:" appears in standard error/output: ```xml <command detect_errors="aggressive"> ... </command> ``` --- ## `stdio` If you need more precision: ```xml <stdio> <exit_code range=":-2" level="warning" description="Low disk space" /> <exit_code range="1:" level="fatal" /> <regex match="Error:" level="fatal" /> </stdio> <command> ... </command> ``` <small>"Warning" level allows to add information to `stderr` without marking the dataset as failed</small> --- ## `help`  ```xml <help><![CDATA[ **What it does** GraPhlAn is a software tool for producing high-quality circular representations of taxonomic and phylogenetic trees. GraPhlAn focuses on concise, integrative, informative, and publication-ready representations of phylogenetically- and taxonomically-driven investigation. For more information, check the `user manual <https://bitbucket.org/nsegata/graphlan/overview>`_. ]]></help> ``` Content should be in [reStructuredText markup format](https://docutils.sourceforge.io/rst.html) --- ## `citations`  ```xml <citations> <citation type="doi">10.1093/bioinformatics/bts611</citation> <citation type="doi">10.1093/nar/gks1219</citation> <citation type="doi">10.1093/nar/gks1005</citation> <citation type="doi">10.1093/bioinformatics/btq461</citation> <citation type="doi">10.1038/nbt.2198</citation> </citations> ``` <small>If no DOI is available, a BibTeX citation can be specified with `type="bibtex"`</small> --- ## Quoting params Always quote `text` and `data` parameters and output `data` in `<command>` ```xml <command><![CDATA[ graphlan.py ... '$input_tree' '$png_output_image' ]]></command> ``` - For security reasons - Paths may contain spaces - Prefer single quotes over double quotes --- ## Multiple commands Use `&&` to concatenate them ```xml <command><![CDATA[ graphlan.py --format '$format' && echo "Yeah it worked!" ]]></command> ``` <small>The job will exit on the first error encountered.</small> --- ## Param argument Use the `argument` tag when a `param` name reflects the command line argument ```xml <param argument="--size" type="integer" value="7" label="..." help="..."/> ``` - It will be appended at the end of the displayed param help - When `argument` is specified and `name` is not, `name` is derived from `argument` by removing the initial dashes and replacing internal dashes with underscores --- ## `section` Use sections to group related parameters  ```xml <section name="advanced" title="Advanced options" expanded="False"> <param argument="--size" type="integer" value="7" label="..." help="..."/> ... </section> ``` ---  > Command-line utilities to assist in building and publishing Galaxy tools. - [Documentation](https://planemo.readthedocs.io/en/latest/) - [Tutorial](https://planemo.readthedocs.io/en/latest/writing_standalone.html) --- ##.image-25[] 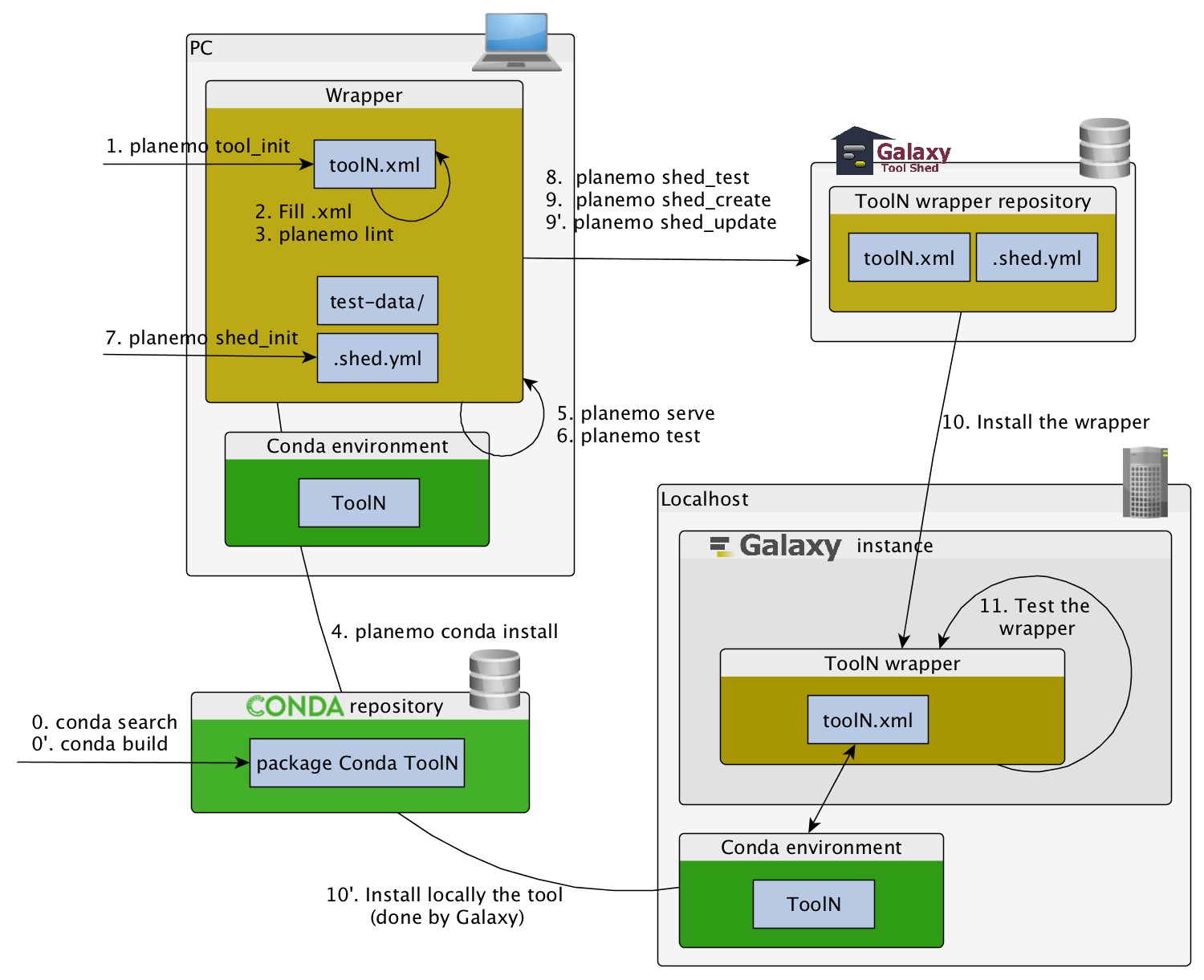 --- ##.image-25[] `planemo tool_init` Creates a skeleton of xml file ```bash $ mkdir new_tool $ cd new_tool $ planemo tool_init --id 'some_short_id' --name 'My super tool' ``` Complicated version: ```bash $ planemo tool_init --id 'samtools_sort' --name 'Samtools sort' \ --description 'order of storing aligned sequences' \ --requirement 'samtools@1.3.1' \ --example_command "samtools sort -o '1_sorted.bam' '1.bam'" \ --example_input 1.bam \ --example_output 1_sorted.bam \ --test_case \ --version_command 'samtools --version | head -1' \ --help_from_command 'samtools sort' \ --doi '10.1093/bioinformatics/btp352' ``` --- class: packed ##.image-25[] `planemo lint` Checks the syntax of a tool ```bash $ planemo lint ``` ```bash Linting tool /opt/galaxy/tools/seqtk_seq.xml Applying linter tests... CHECK .. CHECK: 1 test(s) found. Applying linter output... CHECK .. INFO: 1 outputs found. Applying linter inputs... CHECK .. INFO: Found 1 input parameters. Applying linter help... CHECK .. CHECK: Tool contains help section. .. CHECK: Help contains valid reStructuredText. Applying linter general... CHECK .. CHECK: Tool defines a version [0.1.0]. .. CHECK: Tool defines a name [Convert to FASTA (seqtk)]. .. CHECK: Tool defines an id [seqtk_seq]. Applying linter command... CHECK .. INFO: Tool contains a command. Applying linter citations... CHECK .. CHECK: Found 1 likely valid citations. ``` --- ##.image-25[] `planemo serve` View your new tool in a local Galaxy instance ```bash $ planemo serve ``` Open http://127.0.0.1:9090/ in your web browser to view your new tool --- ##.image-25[] Building Galaxy Tools 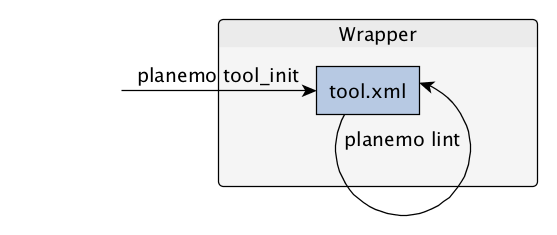 --- # Functional tests --- ## Functional tests - Functional testing is a quality assurance (QA) process. - The tests comfort developers and users that the tools can run across different servers/architectures. And that the latest modifications don't break older features. - Tools are tested by feeding them inputs and parameters and verifying the outputs (typically a diff) --- ## Good practices 1. Build tests using inputs/parameters/outputs 2. Run the tests --> Failed --> Coffee 3. Develop 4. Run the tests --> Failed --> Coffee 5. Develop 6. Run the tests --> Succeed --> Beer --- ## `tests` ```xml <tests> <test> <param name="input_tree" value="input_tree.txt"/> <param name="format" value="png"/> <param name="dpi" value="100"/> <param name="size" value="7"/> <param name="pad" value="2"/> <output name="png_output_image" file="png_image.png" /> </test> </tests> ``` <small>`input_tree.txt` and `png_image.png` must be in the `test-data/` directory</small> --- ## Tool directory tree ``` graphlan/ ├── graphlan.xml ├── graphlan.py └── test-data/ ├── input_tree.txt └── png_image.png ``` --- ## Comparing to an expected result ```xml <output ... compare="diff|re_match|sim_size|contains|re_match_multiline" ... /> ``` ```xml <output name="out_file1" file="cf_maf2fasta_concat.dat" ftype="fasta" /> ``` ```xml <output ... md5="68b329da9893e34099c7d8ad5cb9c940" /> ``` ```xml <output ... lines_diff="4" /> ``` ```xml <output ... compare="sim_size" delta="1000" /> ``` .footnote[[Complete documentation](https://docs.galaxyproject.org/en/latest/dev/schema.html#tool-tests-test-output)] ??? - `diff` is the default - `ftype` also checks the output datatype - With `md5` the test output file doesn't need to be distributed (useful for big output files) - `lines_diff` is useful for tools that output version number, current date, ... - `sim_size` is useful for binary files that vary at each execution (e.g. PDF) --- ## Checking the output content ```xml <output name="out_file1"> <assert_contents> <has_text text="chr7" /> <not_has_text text="chr8" /> <has_text_matching expression="1274\d+53" /> <has_line_matching expression=".*\s+127489808\s+127494553" /> <!-- 	 is XML escape code for tab --> <has_line line="chr7	127471195	127489808" /> <has_n_columns n="3" /> </assert_contents> </output> ``` .footnote[[Complete documentation](https://docs.galaxyproject.org/en/latest/dev/schema.html#tool-tests-test-output-assert-contents)] --- ## Checking tool stdout/stderr ```xml <assert_stdout> <has_text text="Step 1... determine cutoff point" /> <has_text text="Step 2... estimate parameters of null distribution" /> </assert_stdout> ``` .footnote[[Complete documentation](https://docs.galaxyproject.org/en/latest/dev/schema.html#tool-tests-test-output-assert-contents)] --- ## Nested inputs in `test` ```xml <tests> <test> <section name="advanced"> <repeat name="names"> <param name="first" value="Abraham"/> <param name="last" value="Lincoln"/> </repeat> <repeat name="names"> <param name="first" value="Donald"/> <param name="last" value="Trump"/> </repeat> <conditional name="image"> <param name="output_image" value="yes"/> <param name="format" value="png"/> </conditional> </section> ... </test> </tests> ``` --- ##.image-25[] `planemo test` Runs all functional tests ```bash $ planemo test ``` An HTML report is automatically created with logs in case of failing test --- ##.image-25[] Test Galaxy Tools 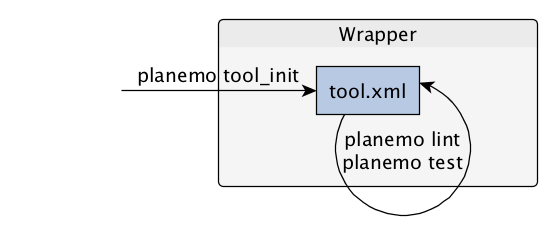 --- # Dependencies --- ## Dependencies How Galaxy will deal with dependencies? 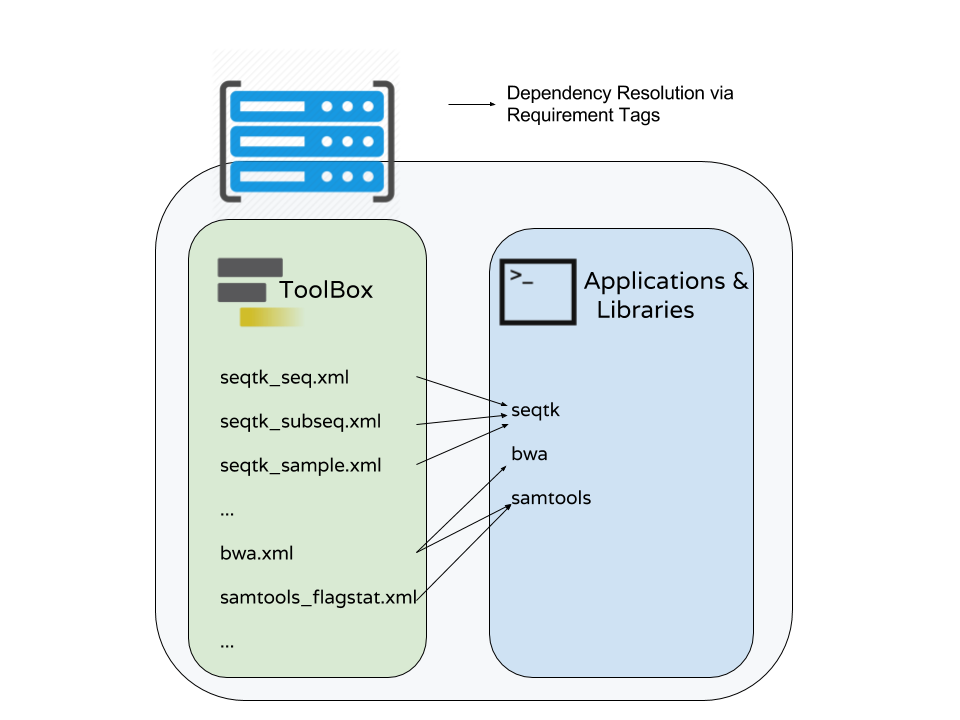 --- ## `requirements` ```xml <requirements> <requirement type="package" version="1.66">biopython</requirement> <requirement type="package" version="1.0.0">graphlan</requirement> </requirements> ``` Local installation using Conda packages --- .image-50[] - Package, dependency and environment manager - Based on recipes describing how to install the software which are then built for their distribution - No compilation at installation: binaries with their dependencies, libraries... - Not restricted to Galaxy See [Tool Dependencies and Conda](../conda/slides.html) --- # Advanced features --- ## `configfiles` A `configfile` creates a text file which can then be used inside the `command` as: - A script or a module - A file needed to run the tool (e.g. a config file) Cheetah code and param/output variables can be used inside `configfile` (like inside `command`). --- class: packed ## `configfiles` ```xml <command><![CDATA[ mb $script_nexus ]]></command> <configfiles> <configfile name="script_nexus"><![CDATA[ set autoclose = yes; execute $input_data; #if str($data_type.type) == "nuc“ lset nst=$data_type.lset_params.lset_Nst; #end if mcmcp ngen=$mcmcp_ngen; mcmc; quit ]]></configfile> </configfiles> ``` ```bash set autoclose = yes; execute dataset_42.dat; lset nst=2 ; mcmcp ngen=100000; mcmc; quit ``` --- ## `macros`  .footnote[[Planemo documentation about macros](https://planemo.readthedocs.io/en/latest/writing_advanced.html#macros-reusable-elements)] --- class: packed ## `macros` > `xml` macros.xml ```xml <macros> <xml name="requirements"> <requirements> <requirement type="package" version="2.5.0">blast</requirement> </requirements> </xml> <xml name="stdio"> <stdio> <exit_code range="1" level="fatal" /> </stdio> </xml> </macros> ``` ncbi_blastn_wrapper.xml ```xml <macros> <import>macros.xml</import> </macros> <expand macro="requirements"/> <expand macro="stdio"/> ``` --- ## `macros` > `token` macros.xml ```xml <macros> <token name="@THREADS@">-num_threads "\${GALAXY_SLOTS:-8}"</token> </macros> ``` ncbi_blastn_wrapper.xml ```xml <command> blastn -query '$query' @THREADS@ [...] </command> ``` --- ## `macros` > `xml` > `yield` macros.xml ```xml <macros> <xml name="requirements"> <requirements> <requirement type="package" version="2.2.0">trinity</requirement> <yield/> </requirements> </xml> </macros> ``` trinity.xml ```xml <expand macro="requirements"> <requirement type="package" version="1.1.2">bowtie</requirement> </expand> ``` --- ## `@TOOL_VERSION@` token ```xml <macros> <token name="@TOOL_VERSION@">1.2</token> </macros> ``` ```xml <tool id="seqtk_seq" name="Convert to FASTA (seqtk)" version="@TOOL_VERSION@+galaxy3"> <requirements> <requirement type="package" version="@TOOL_VERSION@">seqtk</requirement> </requirements> ``` This means: the 3rd revision of the Galaxy tool for Seqtk 1.2 . [Best practice documentation](https://galaxy-iuc-standards.readthedocs.io/en/latest/best_practices/tool_xml.html#tool-versions) --- ## `command` > Reserved variables ```xml <command><![CDATA[ # Email’s numeric ID (id column of galaxy_user table in the database) echo '$__user_id__' # User’s email address echo '$__user_email__' # The galaxy.app.UniverseApplication instance, gives access to all other configuration file variables. # Should be used as a last resort, may go away in future releases. echo '$__app__.config.user_library_import_dir' # Check a dataset type #if $input1.is_of_type('gff'): echo 'input1 type is ${input1.datatype}' #end if ]]></command> ``` .footnote[[Reserved Variables List](https://docs.galaxyproject.org/en/latest/dev/schema.html#reserved-variables)] --- ## Multiple inputs - Mapping over ```xml <param name="..." type="data" format="txt" label="..." help="..." /> ```  Possible to select multiple dataset:  - Useful to launch the same tool on multiple datasets independently - One job per dataset --- ## Multiple inputs - Single execution ```xml <param name="..." type="data" format="txt" multiple="true" label="..." help="..." /> ```  In the command: ```xml <command><![CDATA[ ... #for $input in $inputs --input "$input" #end for ]]></command> ``` One job for all selected dataset --- ## Multiple outputs ```xml <outputs> <data name="output" format="txt"> <discover_datasets pattern="__designation_and_ext__" directory="output_dir" visible="true" /> </data> </outputs> ``` - `__designation_and_ext__`: a predefined regexp, - catches the dataset identifier + the datatype If the output file extension is not present/usable: ```xml <outputs> <data name="output" format="txt"> <discover_datasets pattern="__designation__" format="txt" directory="output_dir" visible="true" /> </data> </outputs> ``` --- ## Dataset collections A dataset collection combines numerous datasets in a single entity that can be manipulated together - `list`: a simple list of datasets - `paired`: a pair of datasets, `forward` and `reverse` for NGS - composite: e.g. `list:paired` for a list of dataset pairs Usage - Useful to launch a workflow on many samples - Sample names are kept along the workflow: `element_identifier` - Galaxy tools are available to manipulate collections --- ## Dataset collections as input Mapping over: ```xml <param name="inputs" type="data" format="bam" label="Input BAM(s)" /> ```  Single execution: - accepted with `multiple="true"` - or accept only collections: ```xml <param name="inputs" type="data_collection" collection_type="list|paired|list:paired|..." format="bam" label="Input BAM(s)" /> ``` ```xml <command><![CDATA[ ... #for $input in $inputs --input '$input' --sample_name '$input.element_identifier' #end for ]]></command> ``` --- ## Dataset collections as output A single paired collection: ```xml <collection name="paired_output" type="paired" label="Split Pair"> <data name="forward" format="txt" /> <data name="reverse" format_source="input1" from_work_dir="reverse.txt" /> </collection> ``` Unknown number of files: ```xml <collection name="output" type="list" label="Unknown number of files"> <discover_datasets pattern="__name_and_ext__" directory="outputs" /> </collection> ``` - `__name_and_ext__`: a predefined regexp, - catches the dataset identifier + the datatype --- ## Use multiple CPUs  ```bash blastn -query foo_bar -num_threads 4 ``` 8 is the default value if not set in destination --- ## Data tables - They list all reference data used by tools - e.g.: Blast databases, BWA indexes, Fasta files - Stored in `.loc` files - Populated by hand or using Data Managers - Data Managers are dedicated kind of Galaxy tools --- ## Using a data table in a tool  --- ## Datatypes - Every Galaxy dataset is associated with a datatype - Datatype can be detected or user specified - Gain of usability .footnote[[Documentation: Adding Datatypes](https://galaxyproject.org/admin/datatypes/adding-datatypes/)] --- # Publishing tools --- ## Contributing to a community Many tools developed by the community on GitHub repositories - [Intergalactic Utilities Commission](https://github.com/galaxyproject/tools-iuc) - [GalaxyP](https://github.com/galaxyproteomics/tools-galaxyp) - ... Added value: - Easier development - Easier contribution for user - Avoid duplications of efforts - Automated tests on each contribution - Automated publishing to ToolShed - Principle of many eyes: if something is visible to many people then, collectively, they are more likely to find errors in it --- ## IUC: Intergalactic Utilities Commission .image-50[] - A team maintaining high quality tools - Establishing and following [best practices](https://galaxy-iuc-standards.readthedocs.io/) for tool development - Open to contributions: bug fixes, new tools, ... https://github.com/galaxyproject/tools-iuc --- ## How should I publish my tool? Adding to an existing GitHub repository (IUC, GalaxyP, ...) - Read the guidelines - Open a pull request - Respond to review comments --- ## How should I publish my tool? Using your own GitHub repository - Reasons: ownership, specific practices, exotic tools - Follow the same structure - Use `.travis.yml` to automate tests and publishing --- ## How should I publish my tool? Using planemo by hand - Ok for few tools - Makes contributing harder [Check out our tutorial to publish to the ToolShed using Planemo](/training-material/topics/dev/tutorials/toolshed/slides.html) --- ## Continuous Integration .image-50[] .image-50[] .image-50[] .image-50[] --- ## Continuous Integration - Create a Pull Request on a GitHub repository - Tests are automatically run on Travis - Other contributors review your tool - The Pull Request is accepted when all lights are green - The tool is automatically uploaded to the ToolShed --- ## GitHub side  --- ## Travis CI side  --- ## ToolShed - Need to create a `.shed.yml` file in the tool directory of the GitHub repository: ```yml categories: [Sequence Analysis] description: Tandem Repeats Finder description long_description: A long long description. name: tandem_repeats_finder_2 owner: gandres ``` ```bash planemo shed_init --name="tandem_repeats_finder_2" --owner="gandres" --description="Tandem Repeats Finder description" --long_description="A long long description." --category="Sequence Analysis" [--remote_repository_url=<URL to .shed.yml on github>] [--homepage_url=<Homepage for tool.>] ``` --- ## Tool suites A tool suite is a group of related tools that can all be installed at once. Defined in `.shed.yml`: implicitly define repositories for each individual tool in the directory and build a suite for those tools. Example: `trinity/.shed.yml` ```yml [...] auto_tool_repositories: name_template: "" description_template: " (from the Trinity tool suite)" suite: name: "suite_trinity" description: Trinity tools to assemble transcript sequences from Illumina RNA-Seq data. ``` --- ## Check ```bash planemo shed_lint --tools --ensure_metadata ``` ```bash Linting repository […]/tandem_repeats_finder Applying linter expansion... CHECK .. INFO: Included files all found. Applying linter tool_dependencies_xsd... CHECK .. INFO: tool_dependencies.xml found and appears to be valid XML Applying linter tool_dependencies_actions... CHECK .. INFO: Parsed tool dependencies. Applying linter repository_dependencies... CHECK .. INFO: No repository_dependencies.xml, skipping. Applying linter shed_yaml... CHECK .. INFO: .shed.yml found and appears to be valid YAML. Applying linter readme... CHECK .. INFO: No README found skipping. +Linting tool […]/tandem_repeats_finder/tandem_repeats_finder_wrapper.xml […] ``` --- .image-50[] .image-50[] .image-50[] --- ## Docker Galaxy Flavors  .footnote[[Docker Galaxy Stable](https://github.com/bgruening/docker-galaxy-stable)] --- ## Docker Galaxy Flavors Dockerfile ```yaml # Galaxy - My own flavour # VERSION 0.1 FROM quay.io/bgruening/galaxy:16.07 MAINTAINER Björn A. Grüning, bjoern.gruening@gmail.com ENV GALAXY_CONFIG_BRAND deepTools # Adding the tool definitions to the container ADD my_tool_list.yml $GALAXY_ROOT/my_tool_list.yml # Install deepTools RUN install-tools $GALAXY_ROOT/my_tool_list.yml ``` --- ## Docker Galaxy Flavors `tool_list.yml` ```yaml api_key: admin galaxy_instance: http://127.0.0.1:8080/ tools: - name: fastqc owner: devteam tool_panel_section_id: cshl_library_information tool_shed_url: https://toolshed.g2.bx.psu.edu revisions: - '8c650f7f76e9' # v0.62 - 'd2cf2c0c8a11' # v0.63 - name: suite_deeptools owner: bgruening tool_panel_section_label: 'deepTools' tool_shed_url: https://toolshed.g2.bx.psu.edu revisions: - '7f5562625ae2' ``` --- ## Docker Galaxy Flavors RUN ```bash docker run -d -p 8080:80 galaxy-deeptools ``` --- ### <i class="fas fa-key" aria-hidden="true"></i><span class="visually-hidden">keypoints</span> Key points - Galaxy Tool Syntax - Use Planemo - Use Conda - Use GitHub - Use Travis - No more excuse to develop crappy Galaxy tools --- ### <i class="fas fa-graduation-cap" aria-hidden="true"></i><span class="visually-hidden">curriculum</span> Do you want to extend your knowledge? Follow one of our recommended follow-up trainings: - [Development in Galaxy](/training-material/topics/dev) - Tool Dependencies and Conda: [<i class="fab fa-slideshare" aria-hidden="true"></i><span class="visually-hidden">slides</span> slides](/training-material/topics/dev/tutorials/conda/slides.html) --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> Authors: <a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-shiltemann"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/shiltemann?s=27" alt="Avatar">Saskia Hiltemann</a> <a href="/training-material/hall-of-fame/bebatut/" class="contributor-badge contributor-bebatut"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/bebatut?s=27" alt="Avatar">Bérénice Batut</a> <a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-abretaud"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/abretaud?s=27" alt="Avatar">Anthony Bretaudeau</a> <a href="/training-material/hall-of-fame/jmchilton/" class="contributor-badge contributor-jmchilton"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/jmchilton?s=27" alt="Avatar">John Chilton</a> <a href="/training-material/hall-of-fame/nsoranzo/" class="contributor-badge contributor-nsoranzo"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/nsoranzo?s=27" alt="Avatar">Nicola Soranzo</a> <a href="/training-material/hall-of-fame/bgruening/" class="contributor-badge contributor-bgruening"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/bgruening?s=27" alt="Avatar">Björn Grüning</a> <a href="/training-material/hall-of-fame/lecorguille/" class="contributor-badge contributor-lecorguille"><img src="https://avatars.githubusercontent.com/lecorguille?s=27" alt="Avatar">Gildas Le Corguillé</a> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTN.png" alt="Galaxy Training Network" style="height: 100px;"/> </div> <a rel="license" href="https://creativecommons.org/licenses/by/4.0/"> This material is licensed under the Creative Commons Attribution 4.0 International License</a>.