New Tutorial: VGP assembly pipeline
We are proud to announce that, as result of the collaboration with the Vertebrate Genomes Project (VGP), a new training describing the VGP assembly pipeline (fig. 1) is now available in the Galaxy Training Network. The Vertebrate Genomes Project aims to generate high-quality, near-error-free, gap-free, chromosome-level, haplotype-phased, annotated reference genome assemblies for every vertebrate species.
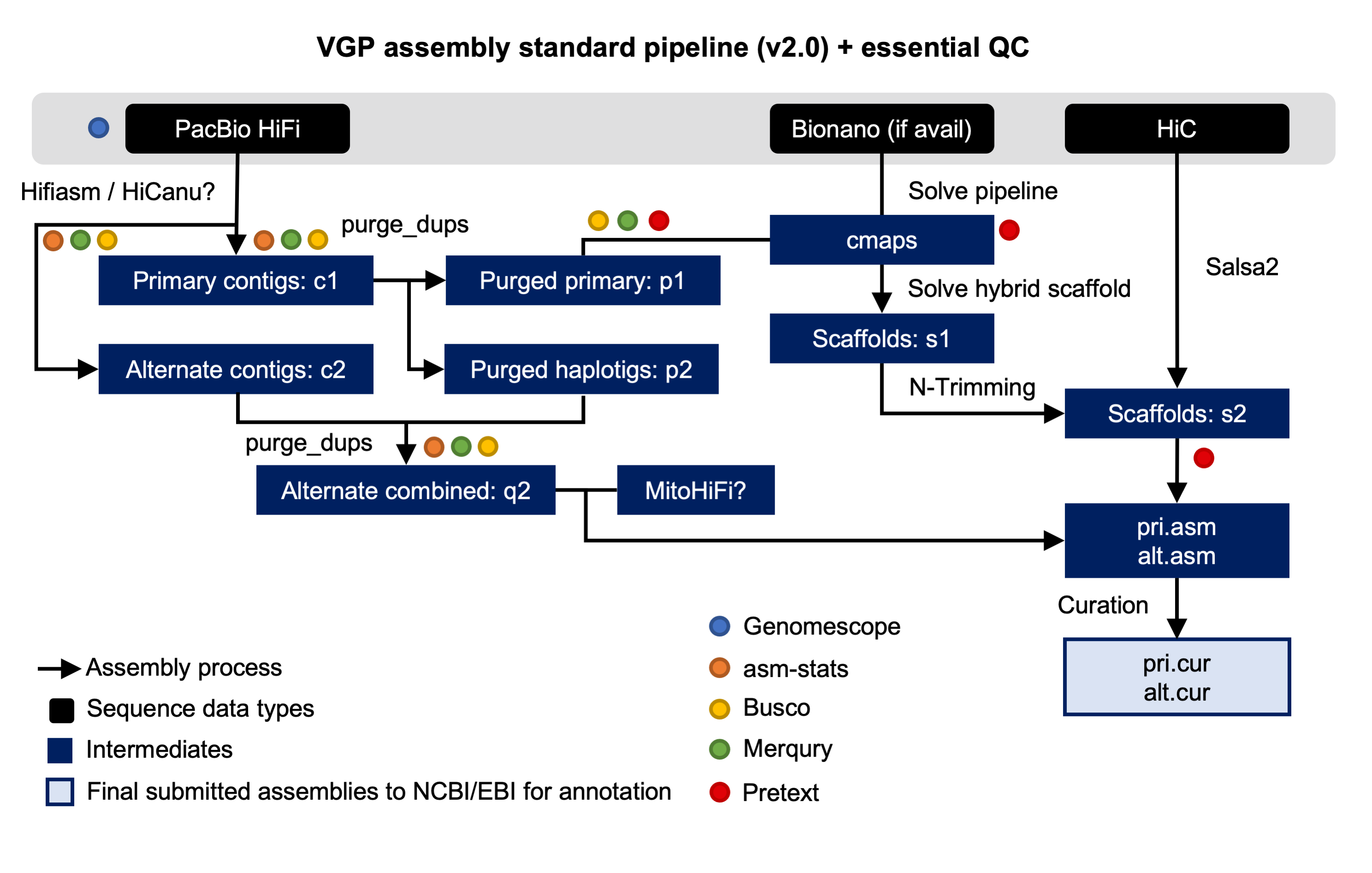
Figure 1. VPG Pipeline 2.0. The pipeline starts with assembly of the HiFi reads into contigs, yielding the primary and alternate assemblies. Then, duplicated and erroneously assigned contigs will be removed by using purge_dups. Finally, Bionano optical maps and HiC data are used to generate a scaffolded primary assembly.
The tutorial organized in four sections: genome profile, HiFi phased assembly, post-assembly pocessing and hybrid scaffolding. During the genome profiling stage, diverse tools based on the analsys of k-mer frequencies are used for infering the properties of the genome. After that, a draft assembly is generated by using high accuracy long-read PacBio HiFi reads. In the third stage, the initial assembly is preprocessed for identifying and reassign allelic contigs. Finally, in the last step the assembed contigs are assembled into scaffolds by using two additional technologies: Bionano optical maps and Hi-C data.
You can access the tutorial by clicking on the following link.
