name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/metagenomics" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a class="nav-link" href="https://github.com/galaxyproject/training-material/edit/main/topics/metagenomics/slides/introduction.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTN.png" alt="Galaxy Training Network" class="cover-logo"/> # Introduction to metagenomics <div markdown="0"> <div class="contributors-line"> Authors: <a href="/training-material/hall-of-fame/bebatut/" class="contributor-badge contributor-bebatut"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/bebatut?s=27" alt="Avatar">Bérénice Batut</a> <a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-shiltemann"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/shiltemann?s=27" alt="Avatar">Saskia Hiltemann</a> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"><i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: Jul 9, 2021</div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 6em;"> <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ## Requirements Before diving into this slide deck, we recommend you to have a look at: - [Introduction to Galaxy Analyses](/training-material/topics/introduction) --- ## Metagenomics --- ## Overview - Why study the microbiome? - 2 different approaches - Amplicon sequencing - Shotgun sequencing - Analysis pipelines - Visualisation options --- ## Why study the microbiome? .pull-left[ - Health care research - Humans are full of microorganisms - Skin, gut, oral cavity, nasal cavity, eyes, .. - Affects health, drug efficacy, etc ] .pull-right[ .image-100[ 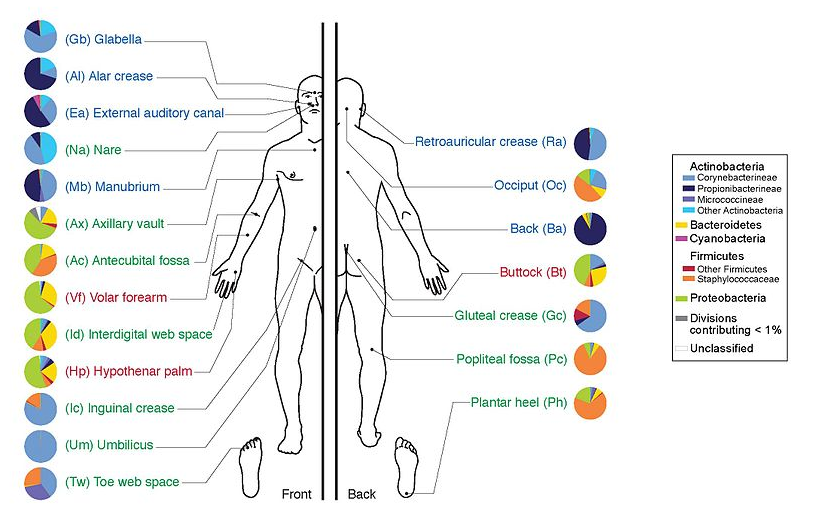 ] ] - Sometimes referred to as your **second genome** - ~10 times more cells than you - ~100 times more genes than you - ~1000s different species --- ## Why study the microbiome? - Environmental studies - Microbes in the soil affect plants and animals - Improve agriculture .image-75[ 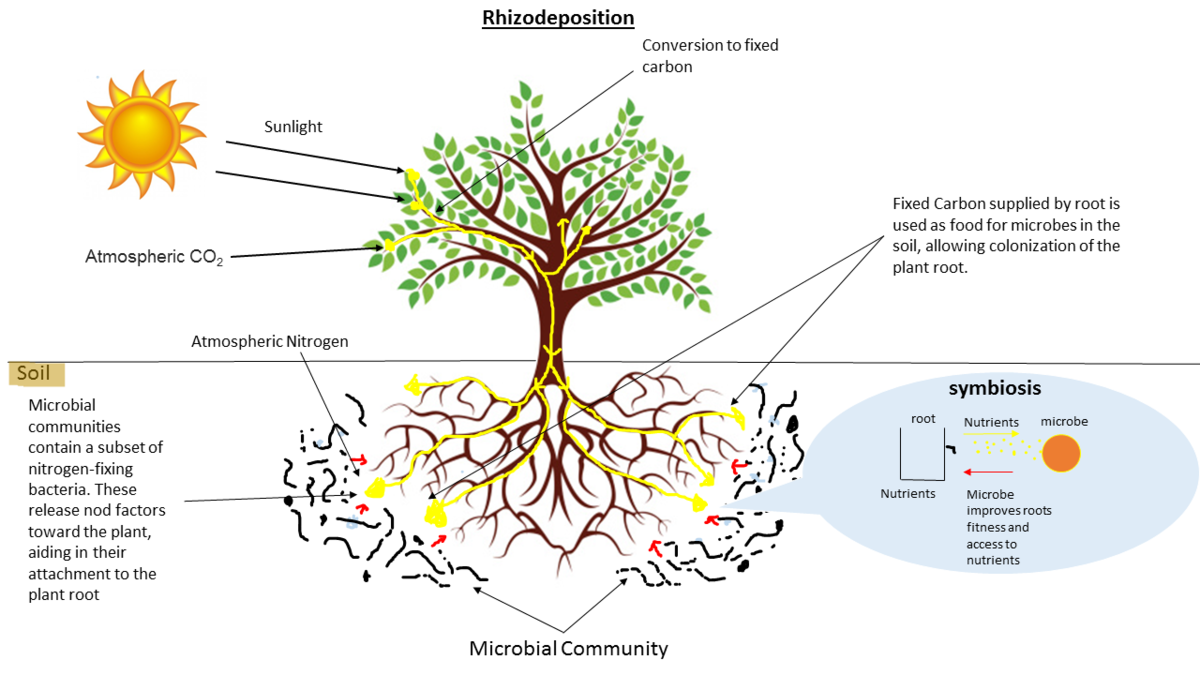 ] --- ## Shotgun vs Amplicon .pull-left[ .image-75[ 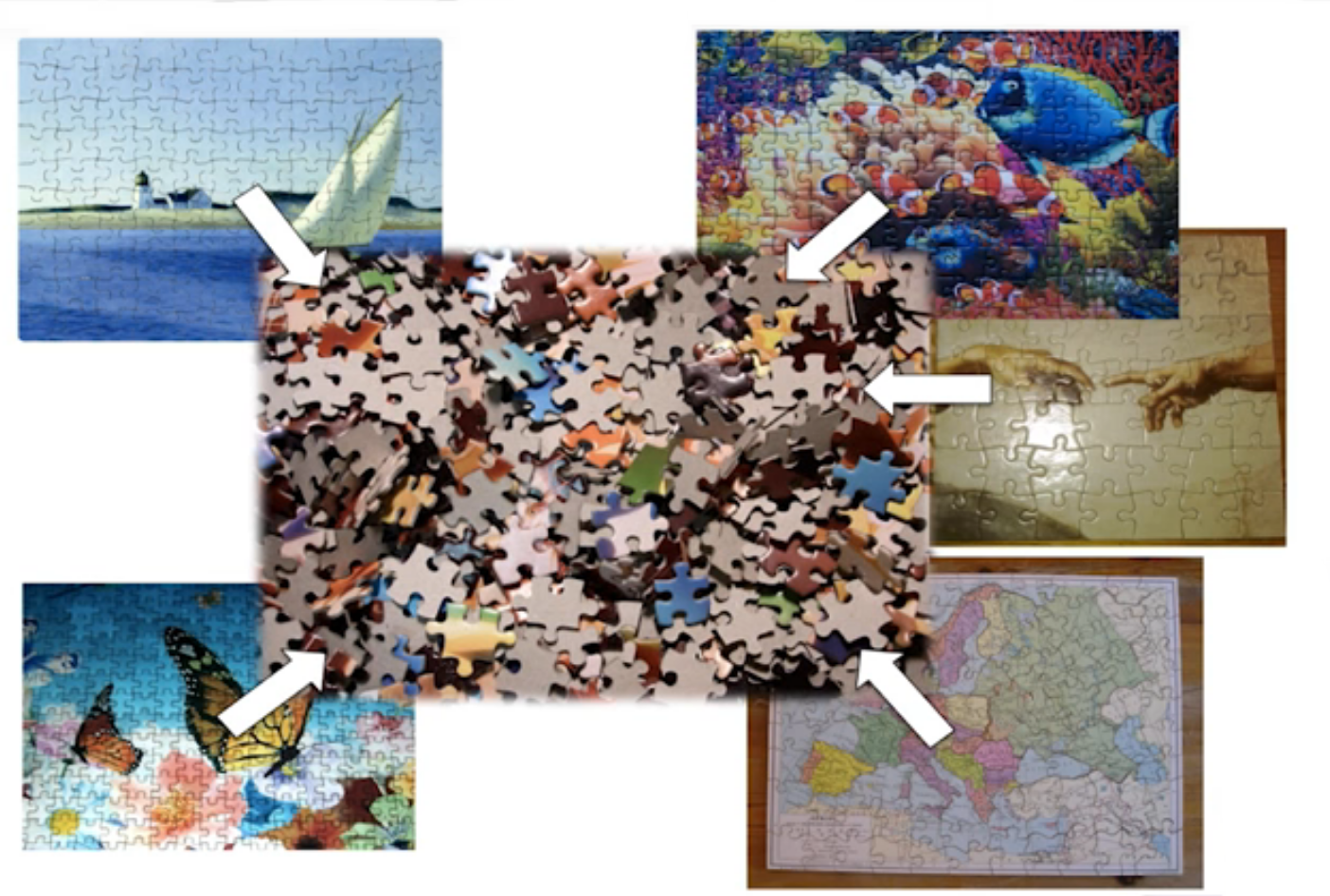 ] **Shotgun Sequencing** - Sequence all DNA - More information - Higher complexity - Higher cost ] .pull-right[ .image-75[  ] **Amplicon Sequencing** - Sequence only specific gene - No functional information - Less complex to analyse - Cheaper ] ??? puzzle analogy: think of each organism as a jigsaw puzzle - shotgun: we considers all pieces (DNA) from all puzzles (organisms); we throw the pieces in a big pile, and need to reconstruct each individual puzzle from this. It is more complex, but you get full details (functional information) about the picture on each puzzle. - amplicon: we only look at the corner pieces, these are easy to spot, and all puzzles have them. It will not tell us all the details (e.g. functional information) of the picture on the puzzle, but might be enough to tell us whether it is a landscape or a portrait or abstract art for instance (taxonomy). --- ## Amplicon - Targetted approach (e.g. 16S/18S rRNA gene) - Amplifies bacteria, not host or environmental fungi, plants 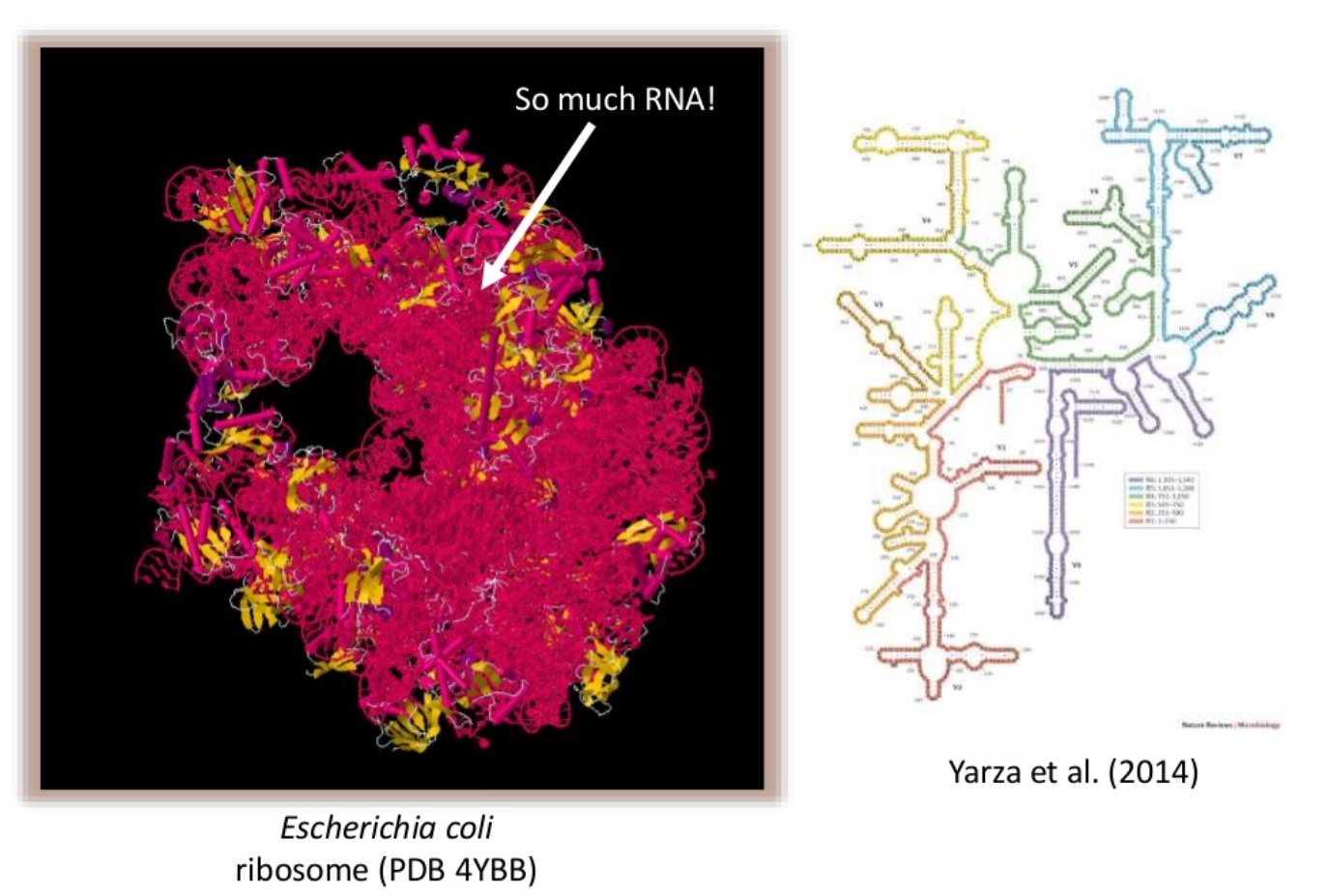 --- ## Amplicon - Highly conserved gene: easy to target across all bacteria - With variable regions: distinguish between genus 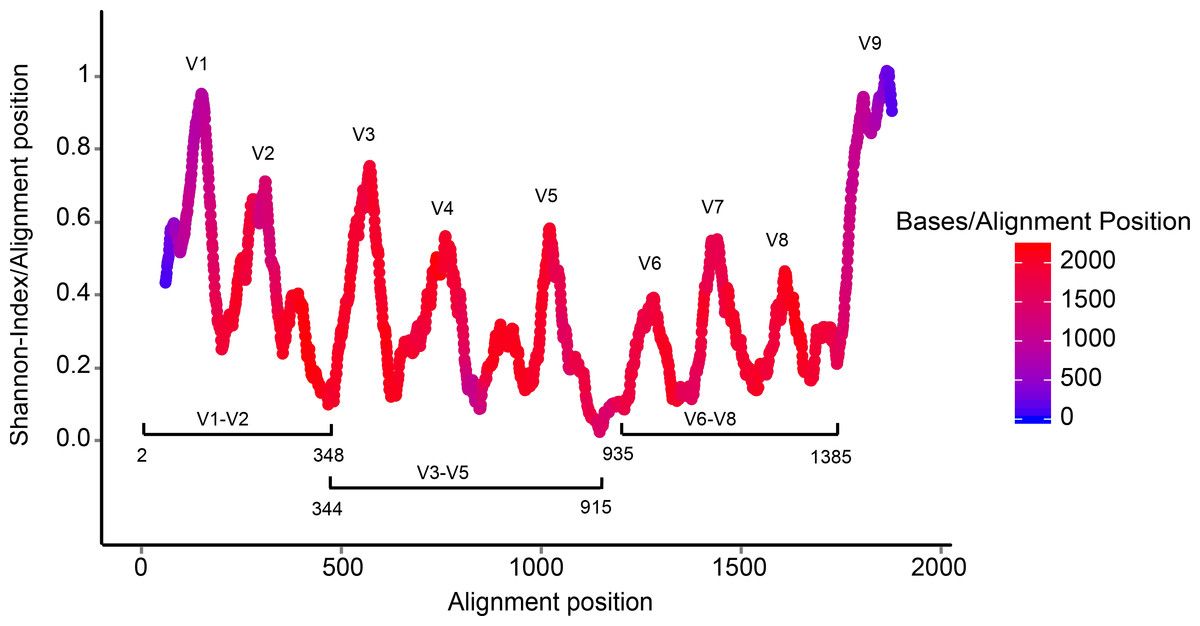 ??? V1, V2 etc are the variable regions --- ## Amplicon - Pros - Well-established - Inexpensive ($50-$100/sample) - Cons - V-region choice can bias results - Is based on a very well conserved gene, making it hard to resolve species and strains ??? 18S gene also amplifies archaea --- ## Shotgun metagenomics - Aims to sequence the "whole" metagenome - Pros: - Not biased by amplicon primer set - Not limited by conservation of the amplicon - Can also provide functional information - Cons: - Environmental contamination, including host - More expensive ($1000+/sample) - Complex data analysis - Requires high performance computing, high memory, high compute capacity --- ## End-to-End  Every step in this process can have serious impact on the results --- ## Bioinformatics  ``` Roche 454 GS: ~ 100.000 reads Illumina MiSeq: ~ 25.000.000 reads Shotgun: ~ ? reads ``` --- ## Analysis pipelines 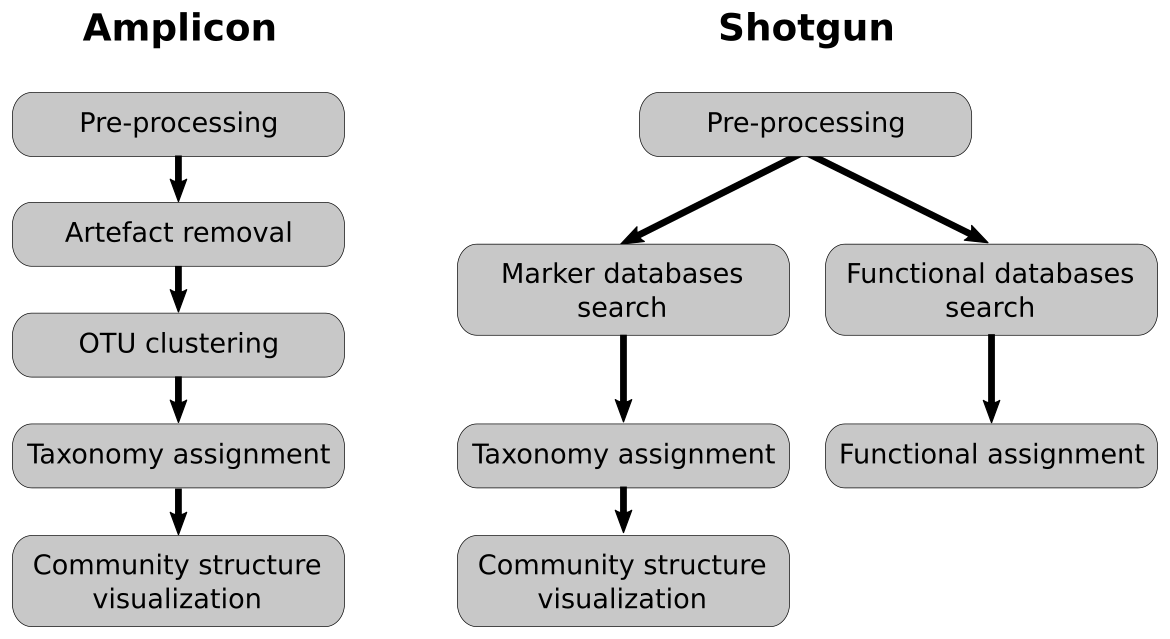 --- ## Pre-processing  - There are a lot of ways to filter and trim your data - Trade-off between quality and amount of information retained --- ## Chimera Removal - During PCR multiple sequences can combine to form a hybrid - Must be removed from your data for better results  --- ## OTU Clustering Cluster on 97% sequence similarity for genus-level differentiation .pull-left[ .image-100[  ]] .pull-right[.image-100[  ]] --- ## Search marker database and taxonomy assignment - Homology with reference databases - Amplicon  - Shotgun: MetaPhlAn2 database - ~1M unique clade-specific marker genes - ~17,000 reference genomes (bacterial and archaeal, viral and eukaryotic) - Accuracy depends on quality and completeness of database used - Databases are inevitable incomplete --- ## Search functional database - Databases to identify gene families - UniRef50 - UniRef90 - Grouping in other functional categories - MetaCyc Reactions - KEGG Orthogroups (KOs) - Pfam domains - Level-4 enzyme commission (EC) categories - EggNOG (including COGs) - Gene Ontology (GO) - Informative GO - Pathway reconstruction --- ## Results: OTU table 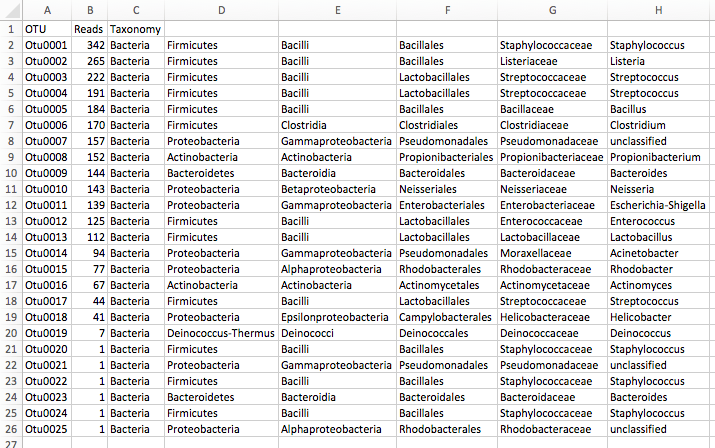 --- ## Results: Visualizations - **Krona** - interactive exploration of sample taxonomy 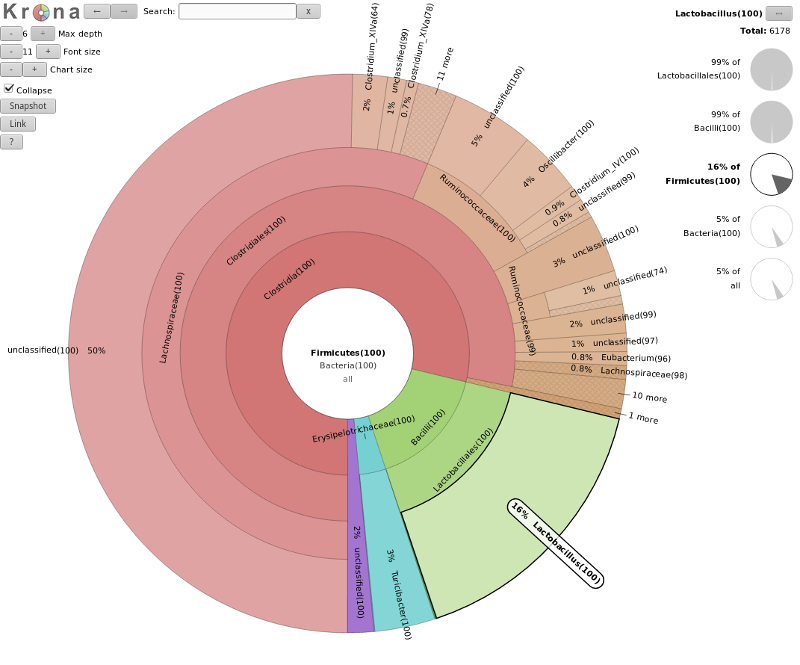 --- ## Results: Visualizations - **Phinch** - BIOM file input - various visualizations - multi-sample data 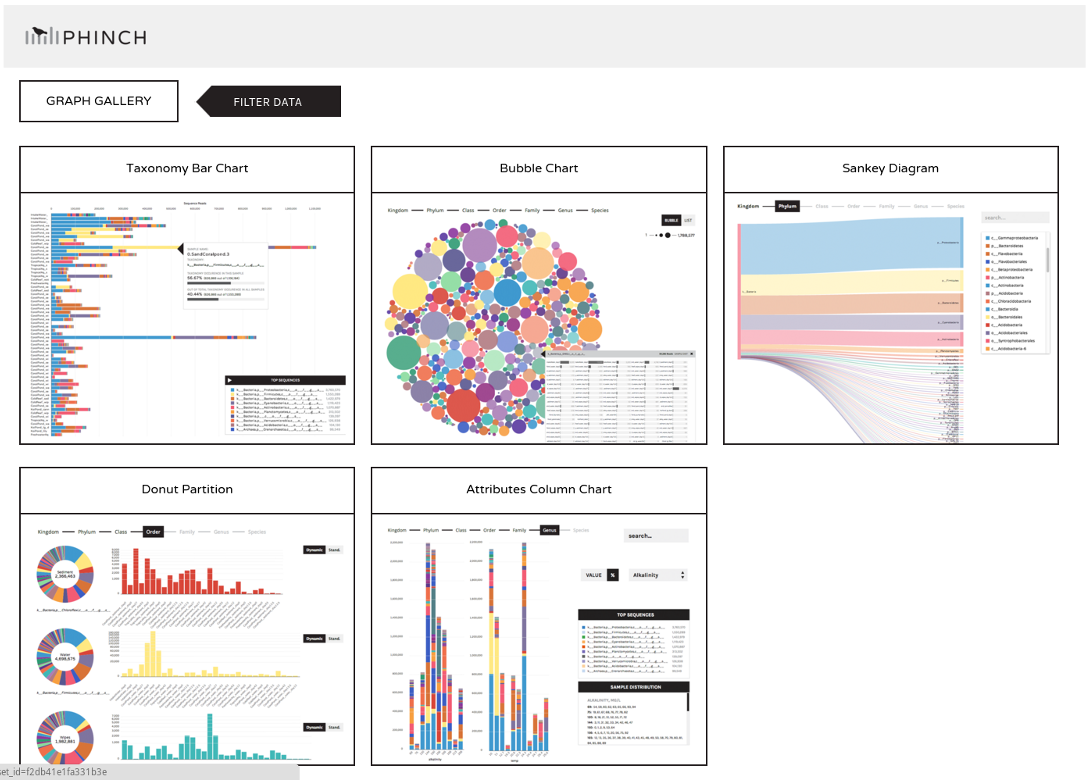 --- ## Related tutorials --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> Authors: <a href="/training-material/hall-of-fame/bebatut/" class="contributor-badge contributor-bebatut"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/bebatut?s=27" alt="Avatar">Bérénice Batut</a> <a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-shiltemann"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/shiltemann?s=27" alt="Avatar">Saskia Hiltemann</a> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTN.png" alt="Galaxy Training Network" style="height: 100px;"/> </div> <a rel="license" href="https://creativecommons.org/licenses/by/4.0/"> This material is licensed under the Creative Commons Attribution 4.0 International License</a>.