name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/dev" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a class="nav-link" href="https://github.com/galaxyproject/training-material/edit/main/topics/dev/tutorials/containers/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTN.png" alt="Galaxy Training Network" class="cover-logo"/> # Tool Dependencies and Containers <div markdown="0"> <div class="contributors-line"> Authors: <a href="/training-material/hall-of-fame/jmchilton/" class="contributor-badge contributor-jmchilton"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/jmchilton?s=27" alt="Avatar">John Chilton</a> <a href="/training-material/hall-of-fame/bgruening/" class="contributor-badge contributor-bgruening"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/bgruening?s=27" alt="Avatar">Björn Grüning</a> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"><i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: Sep 28, 2022</div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 6em;"> <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ## Requirements Before diving into this slide deck, we recommend you to have a look at: - [Development in Galaxy](/training-material/topics/dev) - Tool development and integration into Galaxy: [<i class="fab fa-slideshare" aria-hidden="true"></i><span class="visually-hidden">slides</span> slides](/training-material/topics/dev/tutorials/tool-integration/slides.html) - [<i class="fas fa-laptop" aria-hidden="true"></i><span class="visually-hidden">tutorial</span> hands-on](http://planemo.readthedocs.io/en/latest/writing_standalone.html) --- ### <i class="far fa-question-circle" aria-hidden="true"></i><span class="visually-hidden">question</span> Questions - What are the advantages of running my Galaxy tool inside of a container? - How does Galaxy find a container to run my tool in? - What are BioContainers and how are they related to Galaxy? --- ### <i class="fas fa-bullseye" aria-hidden="true"></i><span class="visually-hidden">objectives</span> Objectives - Explore the differences between containerizing Galaxy and tool execution. - Discuss the advantages of containerizing tools. - Learn to build best practice tools ready to be containerized. --- ## Planemo These slides mirror the section on "Dependencies and Docker" in the [Planemo Documentation](https://planemo.readthedocs.io/en/latest/writing_advanced.html#dependencies-and-docker). --- ## Generic Containers are Good Slide .image-75[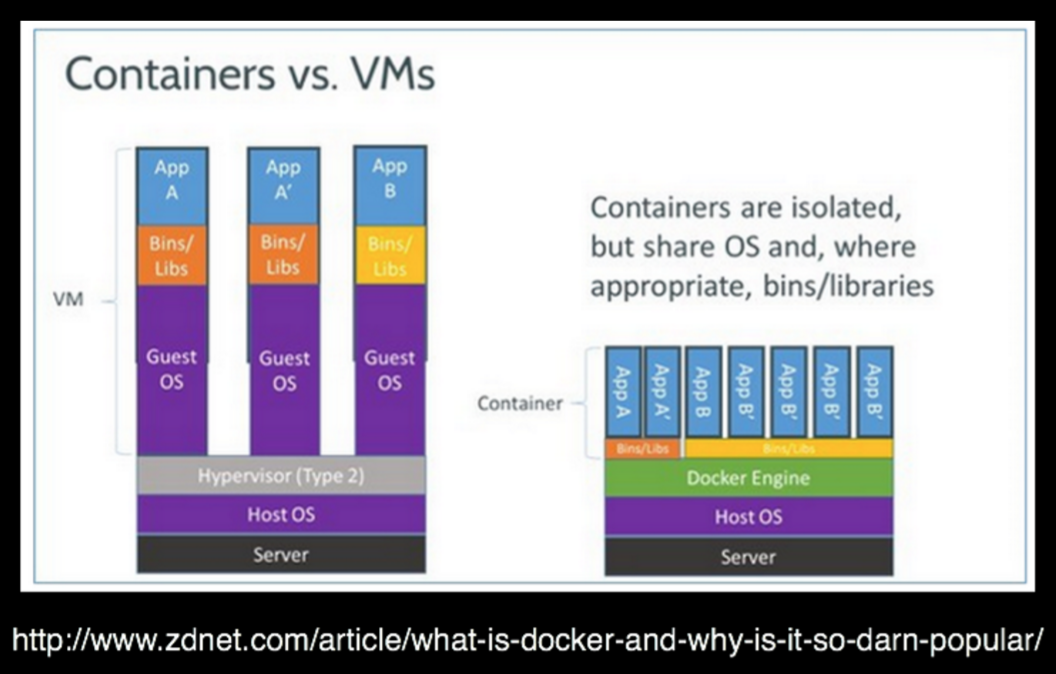] * Isolation and Security\* * Reproducibility * Flexibility\* .footnote[\* the industry is getting there] ??? --- ## Galaxy in Containers? .image-75[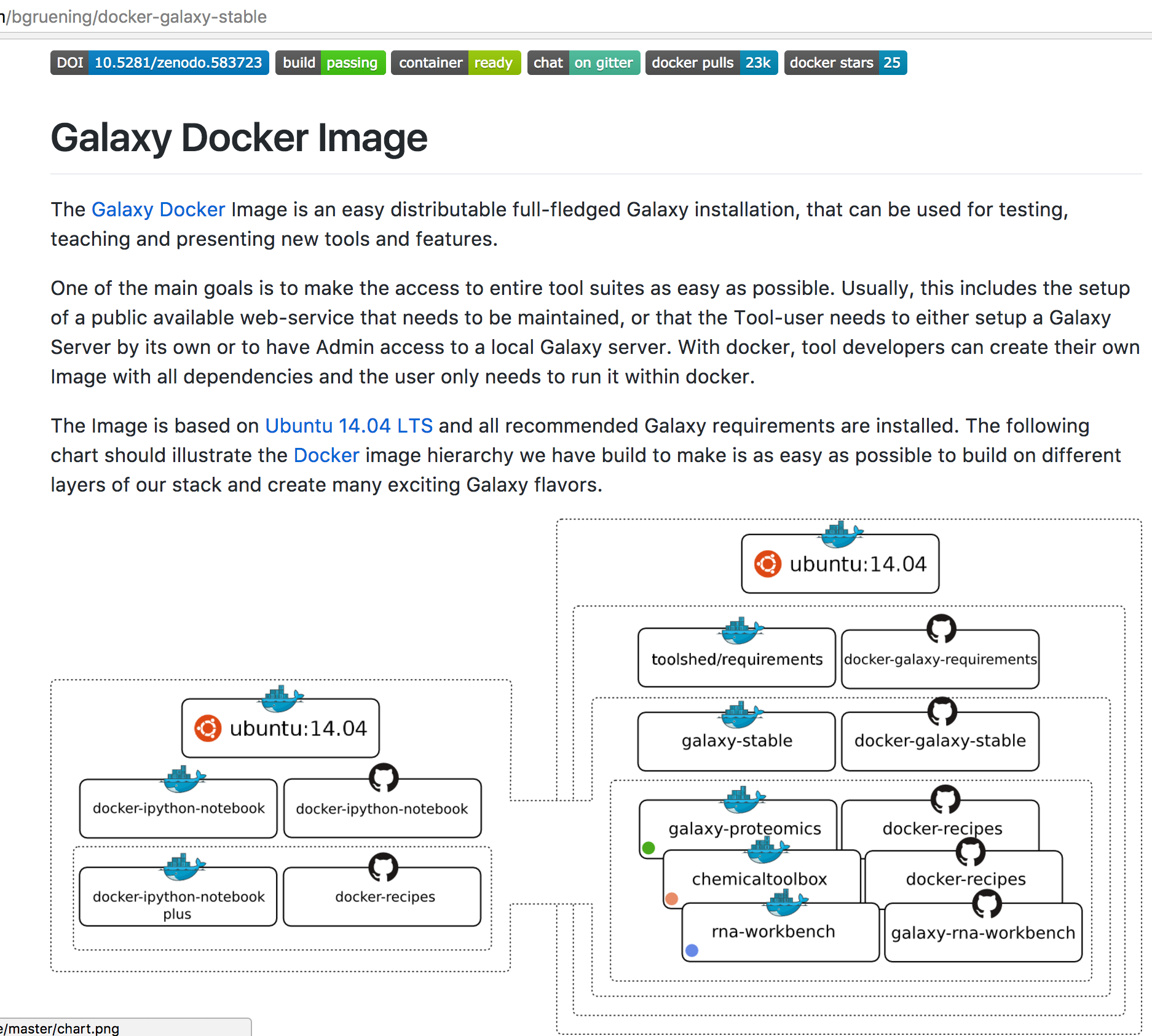] ??? When people think Galaxy and containers I think this project comes to mind. Three years ago Björn created this docker-galaxy-stable project and it has been wildly successful. It has proven to be a useful toolkit for advanced deployment options, an extremely portable way to bring Galaxy to new domains, and a brilliant tool for training users. https://github.com/bgruening/docker-galaxy-stable/commit/27ef7966508958dfec9ce35ff1c5f076ffccf80f Also mention Galaxy KickStart as a way to run Galaxy itself in a container. --- ## Containerizing Galaxy vs Tools We are going to discuss containerizing tool execution instead - executing just jobs in containers. Containers for the particular job's tool. However you deploy Galaxy, including in a container, tool execution can still be containerized. ??? --- class: left ### Containerizing Tools is Still Important * Isolated tool execution. * Isolate file system access. * Added layer of security. * Increased re-computability. * New deployment options - Kubernetes, Mesos Chronos, AWS Batch, etc. --- class: left ### Containerizing Tool Execution Decomposes to basic problems: * <i class="fa fa-square-o" aria-hidden="true"></i> Instruct Galaxy where to find a container for the tool. * <i class="fa fa-square-o" aria-hidden="true"></i> Instruct Galaxy to run the tool in a container. --- class: left ### Configuring Galaxy to Use Containers Just configure the destination. For instance, transform the cluster destination: ``` <destination id="short_fast" runner="slurm"> <param id="nativeSpecification">--time=00:05:00 --nodes=1</param> </destination> ``` as follows: ``` <destination id="short_fast" runner="slurm"> <param id="nativeSpecification">--time=00:05:00 --nodes=1</param> <param id="docker_enabled">true</parma> <param id="docker_sudo">false</param> </destination> ``` That is it! For development, the Planemo flag `--docker` does this for `test`, `serve`, and related commands. --- class: left ### Containerizing Tool Execution #### Decomposes to basic problems * <i class="fa fa-square-o" aria-hidden="true"></i> Instruct Galaxy where to find a container for the tool. * <i class="fa fa-check-square-o" aria-hidden="true"></i> Instruct Galaxy to run the tool in a container. --- class: left ## Explicit Container Dependencies Returning to the seqtk example, lets change the `requirements` from: ``` <requirements> <requirement type="package" version="1.2">seqtk</requirement> </requirements> ``` into ``` <requirements> <container type="docker">quay.io/biocontainers/seqtk:1.2--1</container> <requirement type="package" version="1.2">seqtk</requirement> </requirements> ``` Now run Planemo `test` and `serve` with the `--docker` flag and as a tool developer you are done! --- ### Containerizing Tool Execution #### Decomposes to basic problems * <i class="fa fa-check-square-o" aria-hidden="true"></i> Instruct Galaxy where to find a container for the tool. * <i class="fa fa-check-square-o" aria-hidden="true"></i> Instruct Galaxy to run the tool in a container. We're done... right? ??? --- class: left ## The Problems with Making Docker Explicit * Setting up a `Dockerfile` and publishing a Docker image more process for the tool even though the dependencies have already been completely defined. * An arbitrary Docker image is a blackbox and there is no guarantee Galaxy will execute the same binaries as the Conda requirements. --- class: left ## To Put it Another Way This: ``` <requirements> <requirement type="package" version="1.2">seqtk</requirement> </requirements> ``` should have been sufficient! And the good news is that now it is (mostly)! --- class: left ### .image-10[] BioContainers - The Magic Galaxy can now automatically find or build containers for best practice tools. Planemo will check if such a container has been published with the `--biocontainers` flag to `planemo lint`. ``` $ planemo lint --biocontainers seqtk_seq.xml ... Applying linter biocontainer_registered... CHECK .. INFO: BioContainer best-practice container found [quay.io/biocontainers/seqtk:1.2--1]. ``` ??? But we didn't do anything - where did this magic seqtk container come from. --- ## The Mystery quay.io/biocontainers/seqtk Container 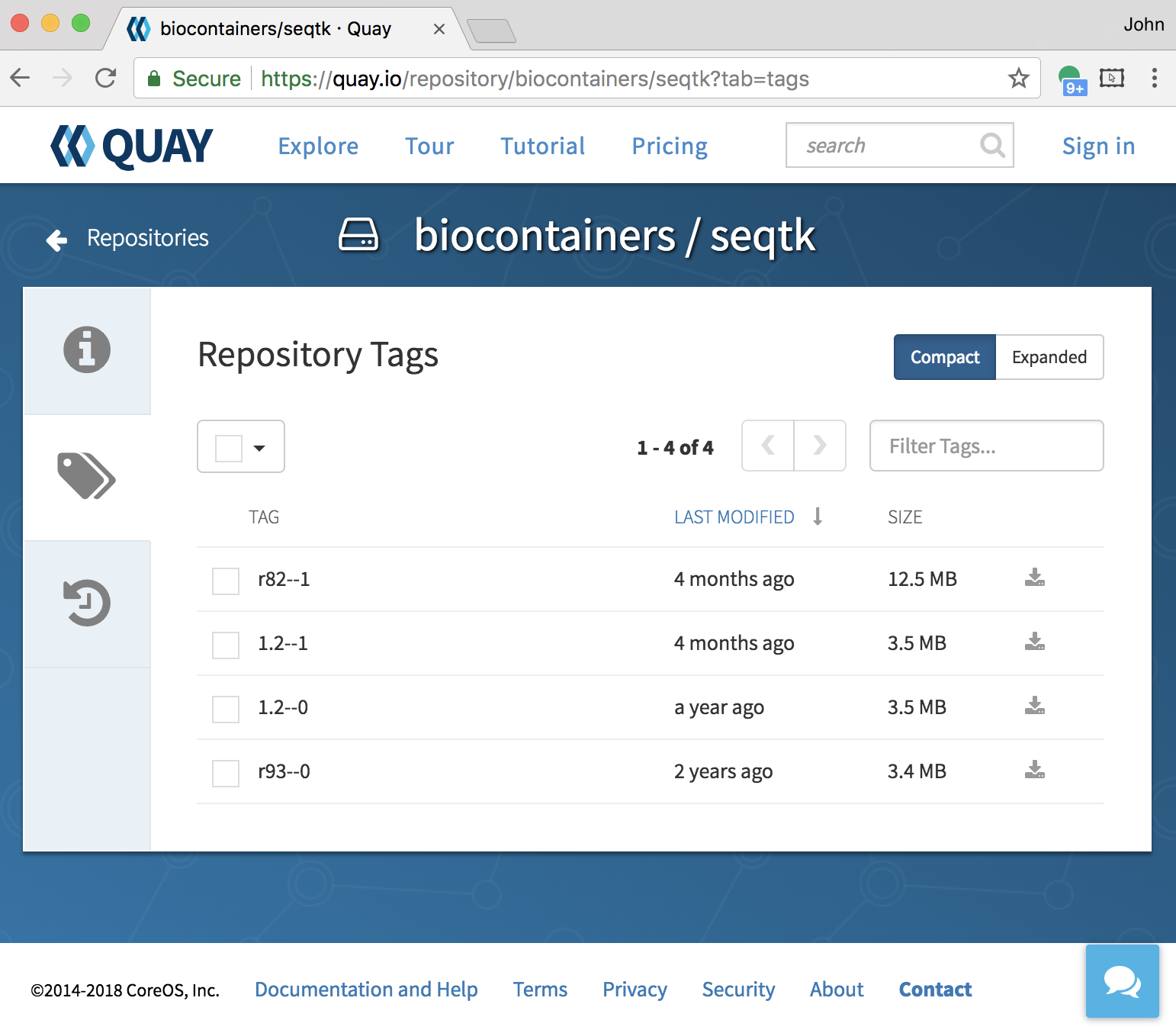 ??? I swear Planemo did not just create that with `tool_init` or `lint`, maybe we should try using it... --- class: left ### .image-05[] BioContainers - Using the Container Run Planemo `test` or `serve` with `--biocontainers` to try mystery container. .reduce70[ ``` $ planemo test --biocontainers seqtk_seq.xml ... [galaxy.tools.actions] Setup for job Job[unflushed,tool_id=seqtk_seq] complete, ready to flush (20.380 ms) [galaxy.tools.actions] Flushed transaction for job Job[id=2,tool_id=seqtk_seq] (15.191 ms) ... [galaxy.tools.deps.containers] Checking with container resolver [ExplicitDockerContainerResolver[]] found description [None] [galaxy.tools.deps.containers] Checking with container resolver [CachedMulledDockerContainerResolver[namespace=None]] found description [None] [galaxy.tools.deps.containers] Checking with container resolver [MulledDockerContainerResolver[namespace=biocontainers]] found description [ContainerDescription[identifier=quay.io/biocontainers/seqtk:1.2--1,type=docker]] [galaxy.jobs.command_factory] Built script [/tmp/tmpw8_UQm/job_working_directory/000/2/tool_script.sh] for tool command [seqtk seq -a '/tmp/tmpw8_UQm/files/000/dataset_1.dat' > '/tmp/tmpw8_UQm/files/000/dataset_2.dat'] ... ok ---------------------------------------------------------------------- XML: /private/tmp/tmpw8_UQm/xunit.xml ---------------------------------------------------------------------- Ran 1 test in 11.926s OK ``` ] The important line here is - `Checking with container resolver [MulledDockerContainerResolver[namespace=biocontainers]] found description [ContainerDescription[identifier=quay.io/biocontainers/seqtk:1.2--1,type=docker]]`. --- ### .image-05[] BioContainers - The Community  ??? BioContainers is a big community of informatics minded individuals trying to work together on building container and Docker based solutions for problems like ours. Previously the project was called BioDocker. We are exploiting and contributing to one aspect of BioContainers, in particular BioConda for Containers. --- ### .image-10[] BioContainers - Bioconda for Containers All Bioconda packages are built into minimal containers. This setup allows the same binaries to be used within the container or on traditional/HPC resources. Without any extra work by tool authors, Galaxy can automatically find or build “the correct” container for a best-practice tool. Over 4,000 containers already published. --- ### .image-05[] BioContainers - Lots of Containers  ??? TODO: Zoom in on the image a bit more ideally. --- ### .image-05[] BioContainers - On Demand Creation Galaxy can be configured to attempt to build containers on demand for containers that haven't been published to the BioContainers namespace on quay.io. These containers will have same names as would be published to quay.io (e.g. `quay.io/biocontainers/seqtk:1.2--0`). --- ### Containerizing Tool Execution #### Decomposes to basic problems * <i class="fa fa-check-square-o" aria-hidden="true"></i><i class="fa fa-check-square-o" aria-hidden="true"></i> Instruct Galaxy where to find a container for the tool. * <i class="fa fa-check-square-o" aria-hidden="true"></i> Instruct Galaxy to run the tool in a container. We're done this time now right? ??? --- ## Revisiting Galaxy Requirements  Many tools have multiple requirements, these need containers also! ??? --- ## An Example The BWA tool is an example of one such tool that has multiple requirements - because samtools is used to sort BAMs after mapping. The Planemo `conda_testing` project is distributed with a simple tool to simulate this containing both `bwa` and `samtools` requirements. .reduce90[ ``` $ planemo project_init --template=conda_testing conda_testing $ cd conda_testing/ $ grep -r require bwa_and_samtools.xml bwa_and_samtools.xml: <requirements> bwa_and_samtools.xml: <requirement type="package" version="0.7.15">bwa</requirement> bwa_and_samtools.xml: <requirement type="package" version="1.3.1">samtools</requirement> bwa_and_samtools.xml: </requirements> ``` ] --- ## Container Hashing Galaxy finds containers based on the names and versions of requirements, so far we have seen the hash for single requirements is just the name and the version. If multiple requirements are present, the hash looks more like a hash. For bwa @ 0.7.15 and samtools @ 1.3.1, Galaxy will look for a container called: .reduce50[`quay.io/biocontainers/mulled-v2-fe8faa35dbf6dc65a0f7f5d4ea12e31a79f73e40:03dc1d2818d9de56938078b8b78b82d967c1f820-0`] --- ## Galaxy Terminology - Mulled To *mull* is to create an environment (either in the Conda sense or globally inside a container) for one or more Conda packages. The result of this is a *mulled* environment. Fixed naming schemes for the resulting environments ensure that different tools with the same set of requirements can reuse a previously created environment or container. If more than one package is included in the resulting environment, a complicated hash is used as this name. --- ## Mulled Hashing Back to the example of bwa at version 0.7.15 and samtools at version 1.3.1, we said the resulting container will be .reduce50[`quay.io/biocontainers/mulled-v2-fe8faa35dbf6dc65a0f7f5d4ea12e31a79f73e40:03dc1d2818d9de56938078b8b78b82d967c1f820-0`] This can be broken into these parts: ``` quay.io/ <namespace> /mulled-v2-<package_hash>:<version_hash>-<build> ``` ??? Explain the different parts. --- ## Exploring Mulled Hashes - by Evgeny Anatskiy  ??? --- class: left ## The Planemo `mull` command (1 / 2) While Galaxy can be configured to auto-build BioContainers as they are needed, the Planemo `mull` command can be used to manually build them for your local Docker host. --- class: reduce70 ## The `mull` command (2 / 2) ``` $ planemo mull bwa_and_samtools.xml /Users/john/.planemo/involucro -v=3 -f /Users/john/workspace/planemo/.venv/lib/python2.7/site-packages/galaxy_lib-17.9.0-py2.7.egg/galaxy/tools/deps/mulled/invfile.lua -set CHANNELS='iuc,bioconda,r,defaults,conda-forge' -set TEST='true' -set TARGETS='samtools=1.3.1,bwa=0.7.15' -set REPO='quay.io/biocontainers/mulled-v2-fe8faa35dbf6dc65a0f7f5d4ea12e31a79f73e40:03dc1d2818d9de56938078b8b78b82d967c1f820' -set BINDS='build/dist:/usr/local/' -set PREINSTALL='conda install --quiet --yes conda=4.3' build /Users/john/.planemo/involucro -v=3 -f /Users/john/workspace/planemo/.venv/lib/python2.7/site-packages/galaxy_lib-17.9.0-py2.7.egg/galaxy/tools/deps/mulled/invfile.lua -set CHANNELS='iuc,bioconda,r,defaults,conda-forge' -set TEST='true' -set TARGETS='samtools=1.3.1,bwa=0.7.15' -set REPO='quay.io/biocontainers/mulled-v2-fe8faa35dbf6dc65a0f7f5d4ea12e31a79f73e40:03dc1d2818d9de56938078b8b78b82d967c1f820' -set BINDS='build/dist:/usr/local/' -set PREINSTALL='conda install --quiet --yes conda=4.3' build DEBU Run file [/Users/john/workspace/planemo/.venv/lib/python2.7/site-packages/galaxy_lib-17.9.0-py2.7.egg/galaxy/tools/deps/mulled/invfile.lua] STEP Run image [continuumio/miniconda:latest] with command [[rm -rf /data/dist]] DEBU Creating container [step-730a02d79e] DEBU Created container [5e4b5f83c455 step-730a02d79e], starting it DEBU Container [5e4b5f83c455 step-730a02d79e] started, waiting for completion DEBU Container [5e4b5f83c455 step-730a02d79e] completed with exit code [0] as expected DEBU Container [5e4b5f83c455 step-730a02d79e] removed STEP Run image [continuumio/miniconda:latest] with command [[/bin/sh -c conda install --quiet --yes conda=4.3 && conda install -c iuc -c bioconda -c r -c defaults -c conda-forge samtools=1.3.1 bwa=0.7.15 -p /usr/local --copy --yes --quiet]] DEBU Creating container [step-e95bf001c8] DEBU Created container [72b9ca0e56f8 step-e95bf001c8], starting it DEBU Container [72b9ca0e56f8 step-e95bf001c8] started, waiting for completion SOUT Fetching package metadata ......... SOUT Solving package specifications: . SOUT SOUT Package plan for installation in environment /opt/conda: SOUT SOUT The following packages will be UPDATED: SOUT SOUT conda: 4.3.11-py27_0 --> 4.3.22-py27_0 SOUT SOUT Fetching package metadata ................. SOUT Solving package specifications: . ... SOUT DEBU Container [72b9ca0e56f8 step-e95bf001c8] completed with exit code [0] as expected DEBU Container [72b9ca0e56f8 step-e95bf001c8] removed STEP Wrap [build/dist] as [quay.io/biocontainers/mulled-v2-fe8faa35dbf6dc65a0f7f5d4ea12e31a79f73e40:03dc1d2818d9de56938078b8b78b82d967c1f820-0] DEBU Creating container [step-6f1c176372] DEBU Packing succeeded ``` This built `quay.io/biocontainers/mulled-v2-fe8faa35dbf6dc65a0f7f5d4ea12e31a79f73e40:03dc1d2818d9de56938078b8b78b82d967c1f820-0`! ??? --- class: reduce70 ## Testing locally `mull`ed containers ``` $ planemo test --galaxy_branch dev --biocontainers bwa_and_samtools.xml ... [galaxy.tools.actions] Handled output named output_2 for tool bwa_and_samtools (17.443 ms) [galaxy.tools.actions] Added output datasets to history (12.935 ms) [galaxy.tools.actions] Verified access to datasets for Job[unflushed,tool_id=bwa_and_samtools] (0.021 ms) [galaxy.tools.actions] Setup for job Job[unflushed,tool_id=bwa_and_samtools] complete, ready to flush (5.755 ms) [galaxy.tools.actions] Flushed transaction for job Job[id=1,tool_id=bwa_and_samtools] (19.582 ms) [galaxy.jobs.handler] (1) Job dispatched [galaxy.tools.deps] Using dependency bwa version 0.7.15 of type conda [galaxy.tools.deps] Using dependency samtools version 1.3.1 of type conda [galaxy.tools.deps] Using dependency bwa version 0.7.15 of type conda [galaxy.tools.deps] Using dependency samtools version 1.3.1 of type conda [galaxy.tools.deps.containers] Checking with container resolver [ExplicitContainerResolver[]] found description [None] [galaxy.tools.deps.containers] Checking with container resolver [CachedMulledContainerResolver[namespace=None]] found description [ContainerDescription[identifier=quay.io/ biocontainers/mulled-v1-01afc412d1f216348d85970ce5f88c984aa443f3:latest,type=docker]] [galaxy.jobs.command_factory] Built script [/tmp/tmpQs0gyp/job_working_directory/000/1/tool_script.sh] for tool command [bwa > /tmp/tmpQs0gyp/files/000/dataset_1.dat 2>&1 ; samtools > /tmp/tmpQs0gyp/files/000/dataset_2.dat 2>&1] [galaxy.tools.deps] Using dependency samtools version None of type conda [galaxy.tools.deps] Using dependency samtools version None of type conda ok ---------------------------------------------------------------------- XML: /private/tmp/tmpQs0gyp/xunit.xml ---------------------------------------------------------------------- Ran 1 test in 7.553s OK [test_driver] Shutting down [test_driver] Shutting down embedded galaxy web server [test_driver] Embedded web server galaxy stopped [test_driver] Stopping application galaxy ... [galaxy.jobs.handler] job handler stop queue stopped Testing complete. HTML report is in "/home/planemo/workspace/planemo/tool_test_output.html". All 1 test(s) executed passed. bwa_and_samtools[0]: passed ``` --- <div id="hands-on-hands-on" class="box-title" aria-label="hands-on box: Hands-on"><i class="fas fa-pencil-alt" aria-hidden="true"></i><span class="visually-hidden"></span> Hands-on: Hands-on</div>  --- <div id="hands-on-hands-on-1" class="box-title" aria-label="hands-on box: Hands-on"><i class="fas fa-pencil-alt" aria-hidden="true"></i><span class="visually-hidden"></span> Hands-on: Hands-on</div> #### The Goal - Use `--biocontainers` to build a container on-demand for a test tool. --- class: left <div id="hands-on-hands-on-2" class="box-title" aria-label="hands-on box: Hands-on"><i class="fas fa-pencil-alt" aria-hidden="true"></i><span class="visually-hidden"></span> Hands-on: Hands-on</div> #### Steps - Run through the bwa_and_samtools.xml test tool and verify container creation. ``` $ planemo project_init --project_template=conda_testing conda_testing $ cd conda_testing/ $ planemo mull bwa_and_samtools.xml $ docker images # verify the container was built $ # use docker run to verify the container has samtools and bwa. $ planemo test --biocontainers bwa_and_samtools.xml ``` --- ### Publishing Multi-Package Containers  ??? We are currently monitoring tools-iuc, tools-devteam, and a half a dozen other repositories daily for package combinations to publish. --- ### Publishing **Your** Multi-Package Containers 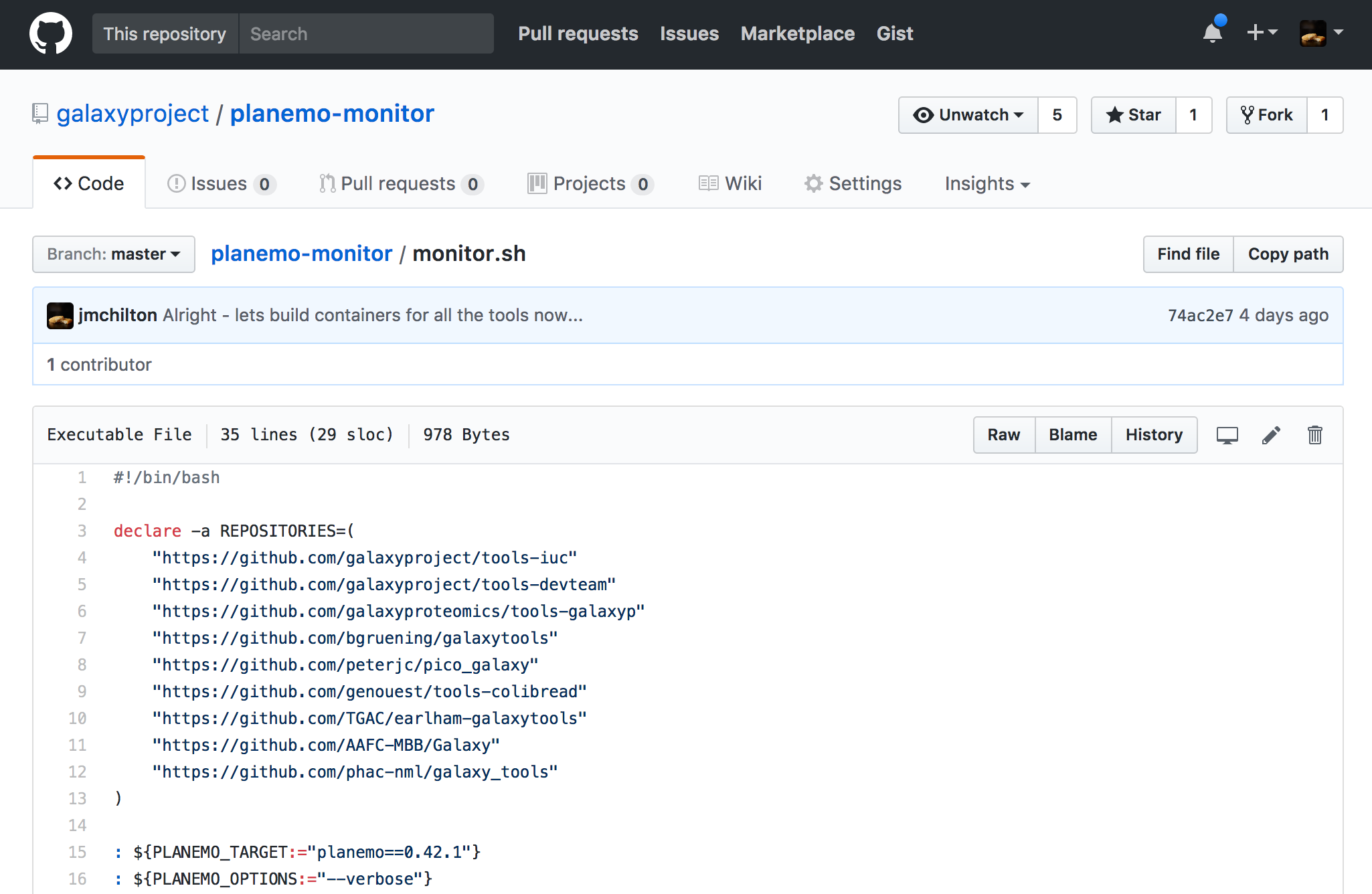 --- ### <i class="fas fa-key" aria-hidden="true"></i><span class="visually-hidden">keypoints</span> Key points - It is becoming easier, more advantageous, and more common for Galaxy admins to run all tools within their own container. - You can explicitly define a container for your tool - but it is easier and more reproducible to let Galaxy find or build one using your tool's best practice requirements. - The Galaxy community will infrastructure to automatically build and/or publish containers for your tool as long as it defines best practice Conda dependencies. - Planemo makes it easy to test your tool inside of containers. --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> Authors: <a href="/training-material/hall-of-fame/jmchilton/" class="contributor-badge contributor-jmchilton"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/jmchilton?s=27" alt="Avatar">John Chilton</a> <a href="/training-material/hall-of-fame/bgruening/" class="contributor-badge contributor-bgruening"><img src="/training-material/assets/images/orcid.png" alt="orcid logo"/><img src="https://avatars.githubusercontent.com/bgruening?s=27" alt="Avatar">Björn Grüning</a> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTN.png" alt="Galaxy Training Network" style="height: 100px;"/> </div> <a rel="license" href="https://creativecommons.org/licenses/by/4.0/"> This material is licensed under the Creative Commons Attribution 4.0 International License</a>.